2JKH
 
 | | Factor Xa - cation inhibitor complex | | Descriptor: | 3-[(3~{a}~{S},4~{R},8~{a}~{S},8~{b}~{R})-4-[5-(5-chloranylthiophen-2-yl)-1,2-oxazol-3-yl]-1,3-bis(oxidanylidene)-4,6,7,8,8~{a},8~{b}-hexahydro-3~{a}~{H}-pyrrolo[3,4-a]pyrrolizin-2-yl]propyl-trimethyl-azanium, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Salonen, L.M, Bucher, C, Banner, D.W, Benz, J, Haap, W, Mary, J.L, Schweizer, W.B, Seiler, P, Kuster, O, Diederich, F. | | Deposit date: | 2008-08-28 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Cation-Pi Interactions at the Active Site of Factor Xa: Dramatic Enhancement Upon Stepwise N-Alkylation of Ammonium Ions.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2HRS
 
 | | Crystal structure of L42H V224H design intermediate for GFP metal ion reporter | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Tubbs, J.L, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2006-07-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Iterative Structure-Based Design of a Green Fluorescent Protein Metal Ion Reporter
To be Published
|
|
2JK7
 
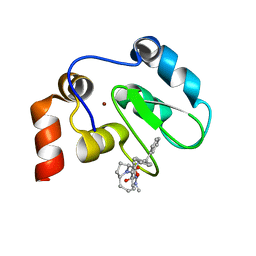 | | XIAP BIR3 bound to a Smac Mimetic | | Descriptor: | (3S,6S,7Z,10AS)-N-(DIPHENYLMETHYL)-6-{[(2S)-2-(METHYLIDENEAMINO)BUTANOYL]AMINO}-5-OXO-1,2,3,5,6,9,10,10A-OCTAHYDROPYRROLO[1,2-A]AZOCINE-3-CARBOXAMIDE, BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 4, ZINC ION | | Authors: | Saito, N.G, Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2008-08-21 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystallographic Studies of Conformationally Constrained Smac Mimetics as Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (Xiap).
J.Med.Chem., 51, 2008
|
|
2JQZ
 
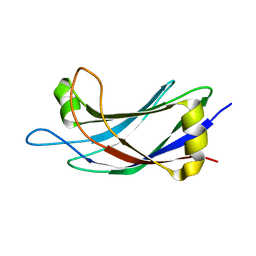 | | Solution Structure of the C2 domain of human Smurf2 | | Descriptor: | E3 ubiquitin-protein ligase SMURF2 | | Authors: | Wiesner, S, Ogunjimi, A.A, Wang, H, Rotin, D, Sicheri, F, Wrana, J.L, Forman-Kay, J.D. | | Deposit date: | 2007-06-15 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of the HECT-Type Ubiquitin Ligase Smurf2 through Its C2 Domain
Cell(Cambridge,Mass.), 130, 2007
|
|
2JJ4
 
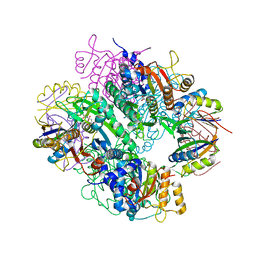 | | The complex of PII and acetylglutamate kinase from Synechococcus elongatus PCC7942 | | Descriptor: | ACETYLGLUTAMATE KINASE, N-ACETYL-L-GLUTAMATE, NITROGEN REGULATORY PROTEIN P-II | | Authors: | Llacer, J.L, Marco-Marin, C, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2007-07-04 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | The Crystal Structure of the Complex of Pii and Acetylglutamate Kinase Reveals How Pii Controls the Storage of Nitrogen as Arginine
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JWN
 
 | |
2K2N
 
 | | Solution structure of a cyanobacterial phytochrome GAF domain in the red light-absorbing ground state | | Descriptor: | PHYCOCYANOBILIN, Sensor protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Ulijasz, A.T, Vierstra, R.D, Markley, J.L. | | Deposit date: | 2008-04-04 | | Release date: | 2008-09-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cyanobacterial phytochrome GAF domain in the red-light-absorbing ground state.
J.Mol.Biol., 383, 2008
|
|
2F1G
 
 | | Cathepsin S in complex with non-covalent 2-(Benzoxazol-2-ylamino)-acetamide | | Descriptor: | Cathepsin S, GLYCEROL, N~2~-1,3-BENZOXAZOL-2-YL-3-CYCLOHEXYL-N-{2-[(4-METHOXYPHENYL)AMINO]ETHYL}-L-ALANINAMIDE | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2005-11-14 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of arylaminoethyl amides as noncovalent inhibitors of cathepsin S. Part 3: Heterocyclic P3.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FA0
 
 | | HMG-CoA synthase from Brassica juncea in complex with HMG-CoA and covalently bound to HMG-CoA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, HMG-CoA synthase | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-06 | | Release date: | 2006-07-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FFT
 
 | | NMR structure of Spinach Thylakoid Soluble Phosphoprotein of 9 kDa in SDS Micelles | | Descriptor: | thylakoid soluble phosphoprotein | | Authors: | Song, J, Carlberg, I, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-12-20 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Micelle-induced folding of spinach thylakoid soluble phosphoprotein of 9 kDa and its functional implications.
Biochemistry, 45, 2006
|
|
2FL7
 
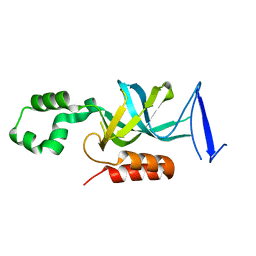 | | S. cerevisiae Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Keck, J.L, Hou, Z, Daner, J.R, Fox, C.A. | | Deposit date: | 2006-01-05 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Sir3 protein bromo adjacent homology (BAH) domain from S. cerevisiae at 1.95 A resolution.
Protein Sci., 15, 2006
|
|
2ETT
 
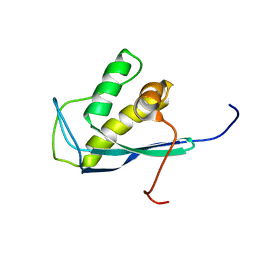 | | Solution Structure of Human Sorting Nexin 22 PX Domain | | Descriptor: | Sorting nexin-22 | | Authors: | Song, J, Zhao, Q, Tyler, R.C, Lee, M.S, Newman, C.L, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human sorting nexin 22.
Protein Sci., 16, 2007
|
|
2JWQ
 
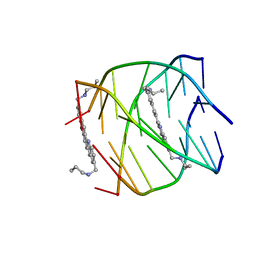 | | G-quadruplex recognition by quinacridines: a SAR, NMR and Biological study | | Descriptor: | DNA (5'-D(*DTP*DTP*DAP*DGP*DGP*DGP*DT)-3'), N,N'-(dibenzo[b,j][1,7]phenanthroline-2,10-diyldimethanediyl)dipropan-1-amine | | Authors: | Hounsou, C, Guittat, L, Monchaud, D, Jourdan, M, Saettel, N, Mergny, J.L, Teulade-Fichou, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | G-Quadruplex Recognition by Quinacridines: a SAR, NMR, and Biological Study
ChemMedChem, 2, 2007
|
|
2K1D
 
 | |
2GJ2
 
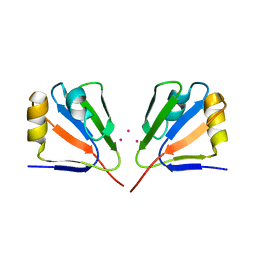 | | Crystal Structure of VP9 from White Spot Syndrome Virus | | Descriptor: | CADMIUM ION, wsv230 | | Authors: | Liu, Y, Wu, J.L, Song, J.X, Sivaraman, J, Hew, C.L. | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of a Novel Nonstructural Protein, VP9, from White Spot Syndrome Virus: Its Structure Reveals a Ferredoxin Fold with Specific Metal Binding Sites
J.Virol., 80, 2006
|
|
2H59
 
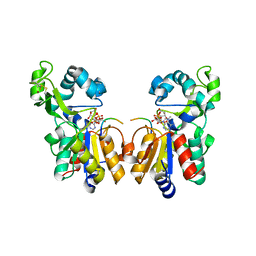 | | Sir2 H116A-deacetylated p53 peptide-3'-o-acetyl ADP ribose | | Descriptor: | (2S,3S,4R,5S)-2-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-4,5-DIHYDROXYTETRAHYDROFURAN-3-YL ACETATE, ADENOSINE-5-DIPHOSPHORIBOSE, Cellular tumor antigen p53, ... | | Authors: | Hoff, K.G, Avalos, J.L, Sens, K, Wolberger, C. | | Deposit date: | 2006-05-25 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD(+) and Acetylated Peptide.
Structure, 14, 2006
|
|
2HFK
 
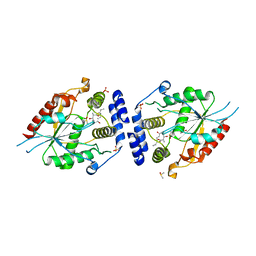 | | Pikromycin thioesterase in complex with product 10-deoxymethynolide | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ETHYL-4-HYDROXY-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Akey, D.L, Kittendorf, J.D, Giraldes, J.W, Fecik, R.A, Sherman, D.H, Smith, J.L. | | Deposit date: | 2006-06-24 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Macrolactonization by the Pikromycin Thioesterase
NAT.CHEM.BIOL., 2, 2006
|
|
2GW6
 
 | |
2GPB
 
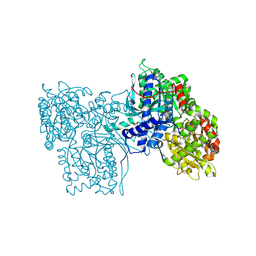 | |
2JER
 
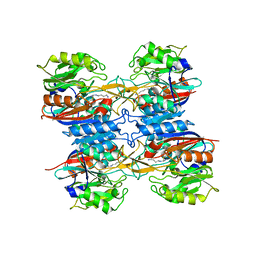 | |
2H4J
 
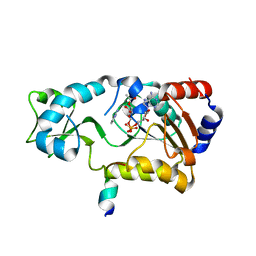 | | Sir2-deacetylated peptide (from enzymatic turnover in crystal) | | Descriptor: | 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, Cellular tumor antigen p53, NAD-dependent deacetylase, ... | | Authors: | Hoff, K.G, Avalos, J.L, Sens, K, Wolberger, C. | | Deposit date: | 2006-05-24 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD(+) and Acetylated Peptide.
Structure, 14, 2006
|
|
2JMU
 
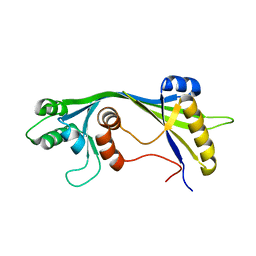 | |
2JO0
 
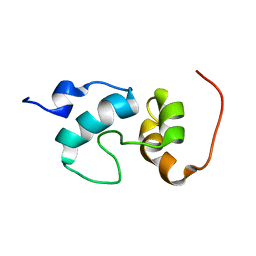 | | The solution structure of the monomeric species of the C terminal domain of the CA protein of HIV-1 | | Descriptor: | Gag-Pol polyprotein | | Authors: | Alcaraz, L.A, del Alamo, M, Barrera, F.N, Mateu, M.G, Neira, J.L. | | Deposit date: | 2007-02-17 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Flexibility in HIV-1 Assembly Subunits: Solution Structure of the Monomeric C-Terminal Domain of the Capsid Protein
Biophys.J., 93, 2007
|
|
2KAN
 
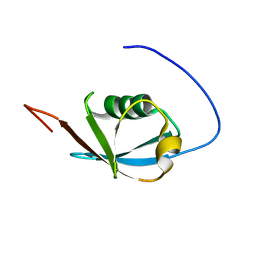 | | Solution NMR structure of ubiquitin-like domain of Arabidopsis thaliana protein At2g32350. Northeast Structural Genomics Consortium target AR3433A | | Descriptor: | uncharacterized protein ar3433a | | Authors: | Eletsky, A, Mills, J.L, Sukumaran, D, Hua, J, Shastry, R, Jiang, M, Ciccosanti, C, Xiao, R, Liu, J, Everret, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-11 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of ubiquitin-like domain of Arabidopsis thaliana protein At2g32350.
To be Published
|
|
2KGQ
 
 | | Refined solution structure of des-pyro Glu brazzein | | Descriptor: | Brazzein | | Authors: | Cornilescu, C.C, Tonelli, M, DeRider, M.L, Markley, J.L, Assadi-Porter, F.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-03-13 | | Release date: | 2009-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of des-pyro Glu brazzein
To be Published
|
|
