3K2O
 
 | | Structure of an oxygenase | | Descriptor: | ACETATE ION, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, CHLORIDE ION, ... | | Authors: | Krojer, T, McDonough, M.A, Clifton, I.J, Mantri, M, Ng, S.S, Pike, A.C.W, Butler, D.S, Webby, C.J, Kochan, G, Bhatia, C, Bray, J.E, Chaikuad, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Schofield, C.J, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the 2-Oxoglutarate- and Fe(II)-Dependent Lysyl Hydroxylase JMJD6.
J.Mol.Biol., 401, 2010
|
|
1QNT
 
 | |
2YPT
 
 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 mutant (E336A) in complex with a synthetic CSIM tetrapeptide from the C-terminus of prelamin A | | Descriptor: | CAAX PRENYL PROTEASE 1 HOMOLOG, PRELAMIN-A/C, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Savitsky, P, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
5TV8
 
 | | A. aeolicus BioW with AMP-CPP and pimelate | | Descriptor: | 6-carboxyhexanoate--CoA ligase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
6BO3
 
 | | Structure Determination of A223, a turret protein in Sulfolobus turreted icosahedral virus, using an iterative hybrid approach | | Descriptor: | Uncharacterized protein | | Authors: | Sendamarai, A.K, Veesler, D, Fu, C.Y, Marceau, C, Larson, E.T, Johnson, J.E, Lawrence, C.M. | | Deposit date: | 2017-11-18 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A223, A STRUCTURAL PROTEIN FROM SULFOLOBUS TURRETED ICOSAHEDRAL VIRUS (STIV)
To Be Published
|
|
3CS1
 
 | | Flagellar Calcium-binding Protein (FCaBP) from T. cruzi | | Descriptor: | Flagellar calcium-binding protein | | Authors: | Ames, J.B, Ladner, J.E, Wingard, J.N, Robinson, H, Fisher, A. | | Deposit date: | 2008-04-08 | | Release date: | 2008-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Membrane Targeting by the Flagellar Calcium-binding Protein (FCaBP), a Myristoylated and Palmitoylated Calcium Sensor in Trypanosoma cruzi.
J.Biol.Chem., 283, 2008
|
|
5TVA
 
 | | A. aeolicus BioW with AMP and CoA | | Descriptor: | 6-carboxyhexanoate--CoA ligase, ADENOSINE MONOPHOSPHATE, COENZYME A | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5VBZ
 
 | | Crystal Structure of Small Molecule Disulfide 2C07 Bound to H-Ras M72C GppNHp | | Descriptor: | 1-(4-methoxyphenyl)-N-(3-sulfanylpropyl)-5-(trifluoromethyl)-1H-pyrazole-4-carboxamide, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Gentile, D.R, Jenkins, M.L, Moss, S.M, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States.
Cell Chem Biol, 24, 2017
|
|
1TZ7
 
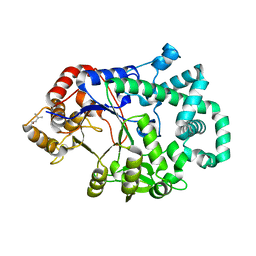 | | Aquifex aeolicus amylomaltase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-alpha-glucanotransferase | | Authors: | Barends, T.R.M, Korf, H, Kaper, T, van der Maarel, M.J.E.C, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2004-07-09 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural influences on product specificity in amylomaltase from Aquifex aeolicus
TO BE PUBLISHED
|
|
5V6I
 
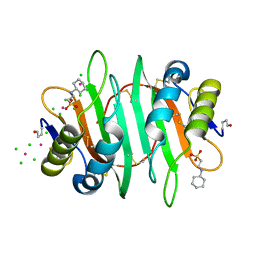 | | Glycan binding protein Y3 from mushroom Coprinus comatus possesses anti-leukemic activity - Pt derivative | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, PLATINUM (II) ION, ... | | Authors: | Li, K, Zhang, P, Gang, Y, Xia, C, Polston, J.E, Li, G, Li, S, Lin, Z, Yang, L.-J, Bruner, S.D, Ding, Y. | | Deposit date: | 2017-03-16 | | Release date: | 2017-08-16 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cytotoxic protein from the mushroom Coprinus comatus possesses a unique mode for glycan binding and specificity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V6J
 
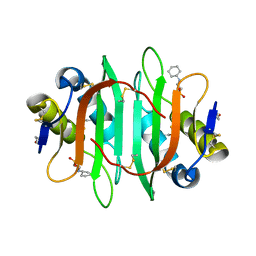 | | Glycan binding protein Y3 from mushroom Coprinus comatus possesses anti-leukemic activity | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, TMV resistance protein Y3 | | Authors: | Li, K, Zhang, P, Gang, Y, Xia, C, Polston, J.E, Li, G, Li, S, Lin, Z, Yang, L.-J, Bruner, S.D, Ding, Y. | | Deposit date: | 2017-03-16 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Cytotoxic protein from the mushroom Coprinus comatus possesses a unique mode for glycan binding and specificity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V6S
 
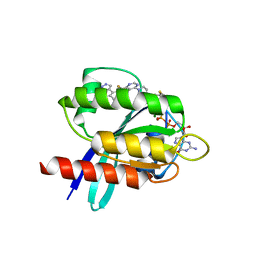 | | Crystal structure of small molecule acrylamide 1 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-8-fluoro-7-(5-methyl-1H-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | McGregor, L.M, Jenkins, M, Kerwin, C, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Expanding the Scope of Electrophiles Capable of Targeting K-Ras Oncogenes.
Biochemistry, 56, 2017
|
|
6AOL
 
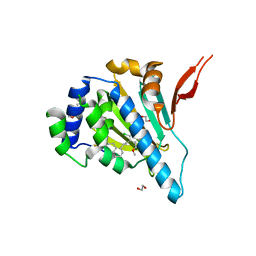 | | Structure of molecular chaperone Grp94 bound to selective inhibitor methyl 3-chloro-2-(2-{2-[(4-fluorophenyl)methyl]phenyl}ethyl)-4,6-dihydroxybenzoate | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endoplasmin, methyl 3-chloro-2-(2-{2-[(4-fluorophenyl)methyl]phenyl}ethyl)-4,6-dihydroxybenzoate | | Authors: | Lieberman, R.L, Huard, D.J.E. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.764 Å) | | Cite: | Second Generation Grp94-Selective Inhibitors Provide Opportunities for the Inhibition of Metastatic Cancer.
Chemistry, 23, 2017
|
|
5T9Y
 
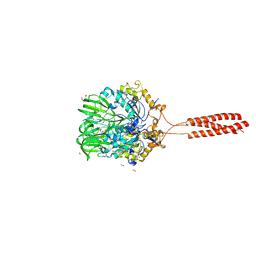 | | Crystal structure of the infectious salmon anemia virus (ISAV) hemagglutinin-esterase protein | | Descriptor: | FORMIC ACID, GLYCEROL, HE protein, ... | | Authors: | Cook, J.D, Sultana, A, Lee, J.E. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-22 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the infectious salmon anemia virus receptor complex illustrates a unique binding strategy for attachment.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3FV8
 
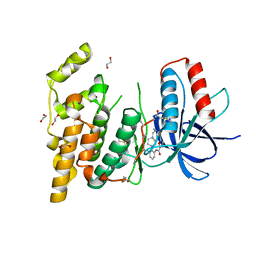 | | JNK3 bound to piperazine amide inhibitor, SR2774. | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(3-chloro-2-(4-(prop-2-ynyl)piperazin-1-yl)phenyl)furan-2-carboxamide, Mitogen-activated protein kinase 10 | | Authors: | Habel, J.E. | | Deposit date: | 2009-01-15 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Synthesis and SAR of piperazine amides as novel c-jun N-terminal kinase (JNK) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7JJL
 
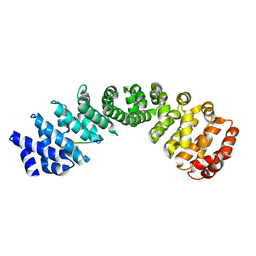 | | Crystal structure of Importin Alpha 3 in complex with human LSD1 NLS | | Descriptor: | Importin subunit alpha-3, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McCuaig, R, Tan, H.Y.A, Hardy, K, Seddiki, N, Ali, S, Dahlstrom, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
5VBE
 
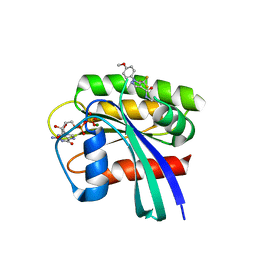 | | Crystal Structure of Small Molecule Disulfide 2C07 Bound to H-Ras M72C GDP | | Descriptor: | 1-(4-methoxyphenyl)-N-(3-sulfanylpropyl)-5-(trifluoromethyl)-1H-pyrazole-4-carboxamide, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gentile, D.R, Jenkins, M.L, Moss, S.M, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States.
Cell Chem Biol, 24, 2017
|
|
2OWC
 
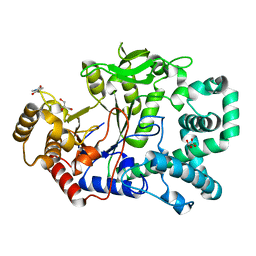 | | Structure of a covalent intermediate in Thermus thermophilus amylomaltase | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-alpha-glucanotransferase, GLYCEROL, ... | | Authors: | Barends, T.R.M, Bultema, J.B, Kaper, T, van der Maarel, M.J.E.C, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2007-02-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Three-way stabilization of the covalent intermediate in amylomaltase, an alpha-amylase-like transglycosylase.
J.Biol.Chem., 282, 2007
|
|
7DFV
 
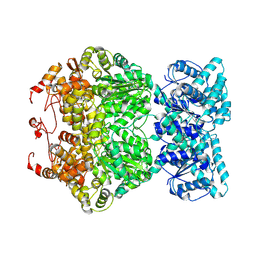 | | Cryo-EM structure of plant NLR RPP1 tetramer core part | | Descriptor: | NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-11-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
5V6V
 
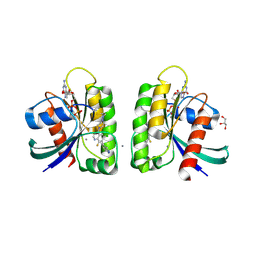 | | Crystal structure of small molecule aziridine 3 covalently bound to K-Ras G12C | | Descriptor: | 3-amino-1-{4-[6-chloro-8-fluoro-7-(5-methyl-1H-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | McGregor, L.M, Jenkins, M, Kerwin, C, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-06-28 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Expanding the Scope of Electrophiles Capable of Targeting K-Ras Oncogenes.
Biochemistry, 56, 2017
|
|
5VBM
 
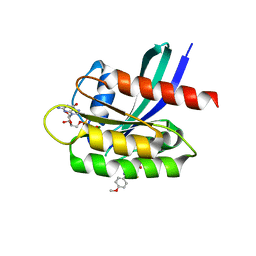 | | Crystal Structure of Small Molecule Disulfide 2C07 Bound to K-Ras Cys Light M72C GDP | | Descriptor: | 1-(4-methoxyphenyl)-N-(3-sulfanylpropyl)-5-(trifluoromethyl)-1H-pyrazole-4-carboxamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gentile, D.R, Jenkins, M.L, Moss, S.M, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States.
Cell Chem Biol, 24, 2017
|
|
3TN8
 
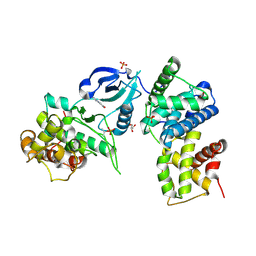 | | CDK9/cyclin T in complex with CAN508 | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Baumli, S, Hole, A.J, Endicott, J.E. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The CDK9 C-helix Exhibits Conformational Plasticity That May Explain the Selectivity of CAN508.
Acs Chem.Biol., 7, 2012
|
|
2OWX
 
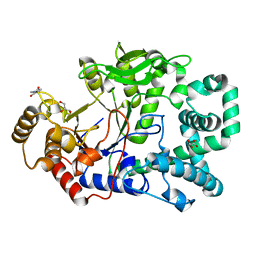 | | THERMUS THERMOPHILUS AMYLOMALTASE AT pH 5.6 | | Descriptor: | 4-alpha-glucanotransferase, GLYCEROL, MALONATE ION | | Authors: | Barends, T.R.M, Kaper, T, Bultema, J.J, Dijkhuizen, L, van der Maarel, J.E.C, Dijkstra, B.W. | | Deposit date: | 2007-02-17 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-way stabilization of the covalent intermediate in amylomaltase, an alpha-amylase-like transglycosylase.
J.Biol.Chem., 282, 2007
|
|
3KVX
 
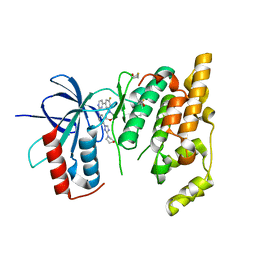 | | JNK3 bound to aminopyrimidine inhibitor, SR-3562 | | Descriptor: | Mitogen-activated protein kinase 10, N-[(2Z)-4-(3-fluoro-5-morpholin-4-ylphenyl)pyrimidin-2(1H)-ylidene]-4-(3-morpholin-4-yl-1H-1,2,4-triazol-1-yl)aniline | | Authors: | Habel, J.E, Laughlin, J.D, LoGrasso, P. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis, Biological Evaluation, X-ray Structure, and Pharmacokinetics of Aminopyrimidine c-jun-N-terminal Kinase (JNK) Inhibitors
J.Med.Chem., 53, 2010
|
|
