4WWW
 
 | | Crystal structure of the E. coli ribosome bound to CEM-101 | | Descriptor: | (3aS,4R,7S,9R,10R,11R,13R,15R,15aR)-1-{4-[4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]butyl}-4-ethyl-7-fluoro-11-methoxy-3a ,7,9,11,13,15-hexamethyl-2,6,8,14-tetraoxotetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-10-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S rRNA, 23S rRNA, ... | | Authors: | Dunkle, J.A, Zhang, W, Cate, J.H.D, Mankin, A.S. | | Deposit date: | 2014-11-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Binding and action of CEM-101, a new fluoroketolide antibiotic that inhibits protein synthesis.
Antimicrob. Agents Chemother., 54, 2010
|
|
5NA4
 
 | | NADH:quinone oxidoreductase (NDH-II) from Staphylococcus aureus - E172S mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH dehydrogenase-like protein SAOUHSC_00878 | | Authors: | Brito, J.A, Athayde, D, Sousa, F.M, Sena, F.V, Pereira, M.M, Archer, M. | | Deposit date: | 2017-02-27 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The key role of glutamate 172 in the mechanism of type II NADH:quinone oxidoreductase of Staphylococcus aureus.
Biochim. Biophys. Acta, 1858, 2017
|
|
8GHS
 
 | | Empty HBV Cp183 capsid with importin-beta, subparticle reconstruction at 2-fold location | | Descriptor: | Capsid protein | | Authors: | Kim, C, Schlicksup, C.J, Hadden-Perilla, J.A, Wang, J.C.-Y, Zlotnick, A. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Hepatitis B virus capsid quasi-6-fold with a trapped C-terminal domain reveals capsid movements associated with domain exit.
J.Biol.Chem., 299, 2023
|
|
8GJ9
 
 | |
8GRJ
 
 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kojima, K, Tsugawa, W, Okuda-Shimazaki, J, Kerrigan, J.A, Sode, K. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone
To Be Published
|
|
8GJA
 
 | | RAD51C-XRCC3 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, RAD51C, ... | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G, Longo, M.A. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
8G2V
 
 | | Cryo-EM structure of recombinant human LECT2 amyloid fibril core | | Descriptor: | Leukocyte cell-derived chemotaxin-2 | | Authors: | Richards, L.S, Flores, M.D, Zink, S, Schibrowsky, N.A, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2023-02-06 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.715 Å) | | Cite: | Cryo-EM structure of a human LECT2 amyloid fibril reveals a network of polar ladders at its core.
Structure, 31, 2023
|
|
8GJ8
 
 | | RAD51C C-terminal domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAD51C | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
4YQ3
 
 | |
4YQA
 
 | |
1PW1
 
 | | Non-Covalent Complex Of Streptomyces R61 DD-Peptidase With A Highly Specific Penicillin | | Descriptor: | (2S,5R,6R)-6-{[(6R)-6-(GLYCYLAMINO)-7-OXIDO-7-OXOHEPTANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLATE, D-alanyl-D-alanine carboxypeptidase, FORMYL GROUP, ... | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-06-30 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
8G0P
 
 | |
1PWC
 
 | | penicilloyl acyl enzyme complex of the Streptomyces R61 DD-peptidase with penicillin G | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, OPEN FORM - PENICILLIN G | | Authors: | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
8G2E
 
 | | PKM2 bound to compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-aminophenyl)methyl]-5-methyl-7-[methyl(oxidanyl)-$l^{3}-sulfanyl]pyridazino[4,5-b]indol-4-one, MAGNESIUM ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Development of Novel Small-Molecule Activators of Pyruvate Kinase Muscle Isozyme 2, PKM2, to Reduce Photoreceptor Apoptosis.
Pharmaceuticals, 16, 2023
|
|
5MF0
 
 | | Crystal structure of Smad4-MH1 bound to the GGCCG site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*GP*CP*CP*CP*GP*T)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5NM3
 
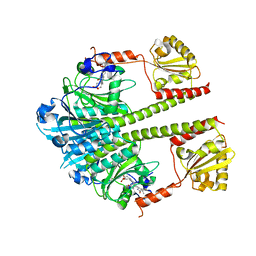 | | Deinococcus radiodurans BphP PAS-GAF-PHY Y263F mutant, pre-illuminated | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Takala, H, Westehoff, S, Ihalainen, J.A. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | On the (un)coupling of the chromophore, tongue interactions, and overall conformation in a bacterial phytochrome.
J. Biol. Chem., 293, 2018
|
|
8FAK
 
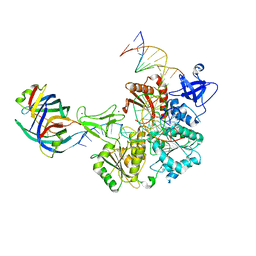 | | DNA replication fork binding triggers structural changes in the PriA DNA helicase that regulate the PriA-PriB replication restart pathway in E. coli | | Descriptor: | DNA (5'-D(P*CP*AP*GP*AP*CP*TP*CP*AP*TP*TP*TP*AP*GP*CP*CP*CP*TP*TP*AP*TP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*AP*TP*AP*AP*GP*GP*GP*CP*TP*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3'), ... | | Authors: | Duckworth, A.T, Ducos, P.L, McMillan, S.D, Satyshur, K.A, Blumenthal, K.H, Deorio, H.R, Larson, J.A, Sandler, S.J, Grant, T, Keck, J.L. | | Deposit date: | 2022-11-28 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Replication fork binding triggers structural changes in the PriA helicase that govern DNA replication restart in E. coli.
Nat Commun, 14, 2023
|
|
1YSO
 
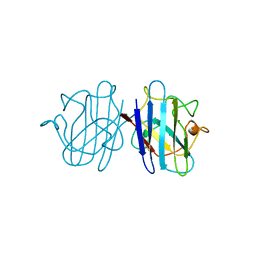 | | YEAST CU, ZN SUPEROXIDE DISMUTASE WITH THE REDUCED BRIDGE BROKEN | | Descriptor: | COPPER (I) ION, YEAST CU, ZN SUPEROXIDE DISMUTASE, ... | | Authors: | Parge, H.E, Crane, B.R, Tsang, J, Tainer, J.A. | | Deposit date: | 1995-12-21 | | Release date: | 1996-06-10 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unusual trigonal-planar copper configuration revealed in the atomic structure of yeast copper-zinc superoxide dismutase.
Biochemistry, 35, 1996
|
|
5NXD
 
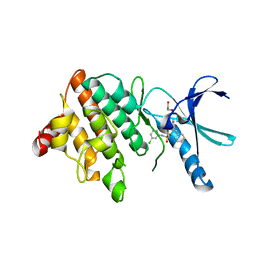 | | LIM Domain Kinase 2 (LIMK2) In Complex With TH-300 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-chlorophenyl)sulfamoyl]-~{N}-(phenylmethyl)-~{N}-propyl-benzamide, LIM domain kinase 2 | | Authors: | Mathea, S, Salah, E, Hanke, T, Newman, J.A, Oerum, S, Wang, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Knapp, S. | | Deposit date: | 2017-05-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | LIM Domain Kinase 2 (LIMK2)In Complex With TH-300
To Be Published
|
|
1CWP
 
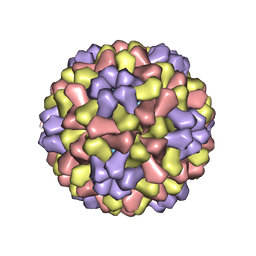 | | STRUCTURES OF THE NATIVE AND SWOLLEN FORMS OF COWPEA CHLOROTIC MOTTLE VIRUS DETERMINED BY X-RAY CRYSTALLOGRAPHY AND CRYO-ELECTRON MICROSCOPY | | Descriptor: | Coat protein, RNA (5'-R(*AP*U)-3'), RNA (5'-R(*AP*UP*AP*U)-3') | | Authors: | Speir, J.A, Johnson, J.E, Munshi, S, Wang, G, Timothy, S, Baker, T.S. | | Deposit date: | 1995-05-22 | | Release date: | 1995-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy.
Structure, 3, 1995
|
|
5LRB
 
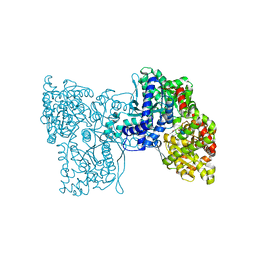 | | Plastidial phosphorylase from Barley in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-1,4 glucan phosphorylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Kruzewicz, K, Palcic, M.M. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development.
PLoS ONE, 12, 2017
|
|
4Z6I
 
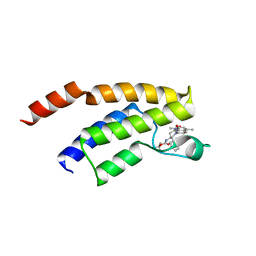 | | Crystal structure of BRD9 bromodomain in complex with a substituted valerolactam quinolone ligand | | Descriptor: | Bromodomain-containing protein 9, tert-butyl [(2R,3S)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxo-2-phenylpiperidin-3-yl]carbamate | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-04-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5M19
 
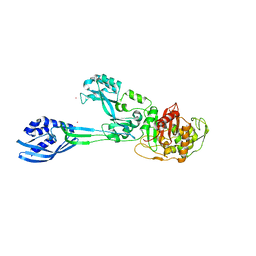 | | Crystal structure of PBP2a from MRSA in the presence of Oxacillin ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
5LR8
 
 | | Structure of plastidial phosphorylase Pho1 from Barley | | Descriptor: | Alpha-1,4 glucan phosphorylase, CITRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Kruzewicz, K, Palcic, M.M. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development.
PLoS ONE, 12, 2017
|
|
5OAU
 
 | |
