7PL2
 
 | |
7PL5
 
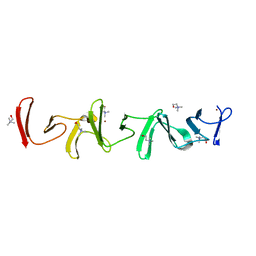 | | Crystal structure of choline-binding module (R1-R9) of LytB from Streptococcus pneumoniae | | Descriptor: | CHOLINE ION, Putative endo-beta-N-acetylglucosaminidase, TRIETHYLENE GLYCOL, ... | | Authors: | Molina, R, Martinez Caballero, S, Hermoso, J.A. | | Deposit date: | 2021-08-28 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis of the final step of cell division in Streptococcus pneumoniae.
Cell Rep, 42, 2023
|
|
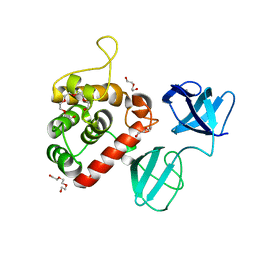
7PJ3
 
 | |
2HBY
 
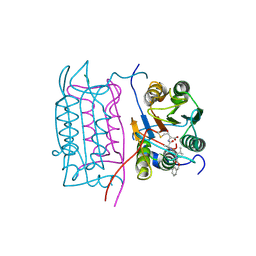 | |
7PL3
 
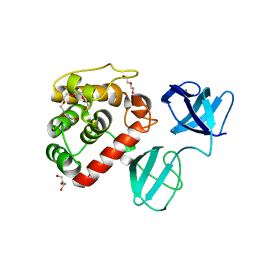 | |
2HDL
 
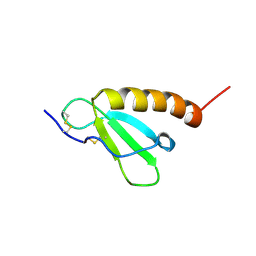 | | Solution structure of Brak/CXCL14 | | Descriptor: | Small inducible cytokine B14 | | Authors: | Peterson, F.C, Thorpe, J.A, Harder, A.G, Volkman, B.F, Schwarze, S.R. | | Deposit date: | 2006-06-20 | | Release date: | 2006-10-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Determinants Involved in the Regulation of CXCL14/BRAK Expression by the 26 S Proteasome.
J.Mol.Biol., 363, 2006
|
|
7POD
 
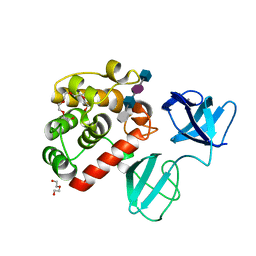 | |
7PPX
 
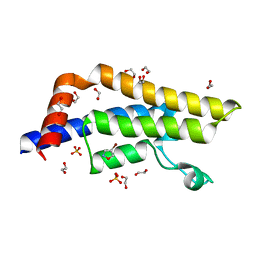 | | ATAD2 in complex with FragLite3 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1,2-oxazole, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Wood, D.J, Ng, Y.M, Heath, R, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
2HI9
 
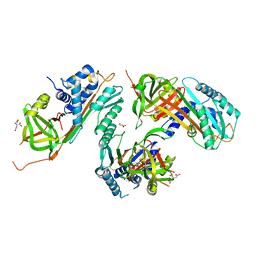 | |
7PMP
 
 | |
2HGD
 
 | |
2HGY
 
 | |
7PQ4
 
 | |
2HG6
 
 | | Solution NMR Structure of Protein PA1123 from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium Target PaT4; Ontario Centre for Structural Proteomics Target PA1123. | | Descriptor: | Hypothetical protein | | Authors: | Lemak, A, Srisailam, S, Yee, A, Lukin, J.A, Orekhov, V.Y, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a hypothetical protein from Pseudomonas aeruginosa (Northeast Structural Genomics Consortium Target: PaT4; Ontario Centre for Structural Proteomics Target: PA1123)
To be Published
|
|
2HI2
 
 | | Crystal structure of native Neisseria gonorrhoeae Type IV pilin at 2.3 Angstroms Resolution | | Descriptor: | Fimbrial protein, HEPTANE-1,2,3-TRIOL, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, ... | | Authors: | Craig, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2006-06-28 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Type IV Pilus Structure by Cryo-Electron Microscopy and Crystallography: Implications for Pilus Assembly and Functions.
Mol.Cell, 23, 2006
|
|
7PTH
 
 | |
7PTJ
 
 | |
7QGI
 
 | | Crystal structure of SARS-CoV-2 NSP14 in the absence of NSP10 | | Descriptor: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-08 | | Release date: | 2022-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
7QGP
 
 | | STK10 (LOK) bound to Macrocycle CKJB51 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-chloranyl-2-methyl-phenyl)methyl]-3-(7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaen-5-yl)urea, CHLORIDE ION, ... | | Authors: | Berger, B.-T, Schroeder, M, Kraemer, A, Schwalm, M.P, Kurz, C.G, Amrhein, J.A, Hanke, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STK10 bound to CKJB51
To Be Published
|
|
7QIF
 
 | | Crystal structure of SARS-CoV-2 NSP14 in complex with 7MeGpppG. | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
2JH5
 
 | | Human Thrombin Hirugen Inhibitor complex | | Descriptor: | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHENESULFONAMIDE, CALCIUM ION, HIRUDIN IIIA, ... | | Authors: | Senger, S, Chan, C, Convery, M.A, Hubbard, J.A, Young, R.J, Shah, G.P, Watson, N.S. | | Deposit date: | 2007-02-20 | | Release date: | 2007-05-08 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sulfonamide-Related Conformational Effects and Their Importance in Structure-Based Design.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7QUE
 
 | | The STK17A (DRAK1) Kinase Domain Bound to CKJB68 | | Descriptor: | Serine/threonine-protein kinase 17A, ~{N}-(phenylmethyl)-7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxamide | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Dederer, V, Kurz, C.G, Amrhein, J.A, Hanke, T, Knapp, S. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Illuminating the Dark: Highly Selective Inhibition of Serine/Threonine Kinase 17A with Pyrazolo[1,5- a ]pyrimidine-Based Macrocycles.
J.Med.Chem., 65, 2022
|
|
2JGO
 
 | | Structure of the arsenated de novo designed peptide Coil Ser L9C | | Descriptor: | ARSENIC, COIL SER L9C, ZINC ION | | Authors: | Touw, D.S, Nordman, C.E, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2007-02-13 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identifying Important Structural Characteristics of Arsenic Resistance Proteins by Using Designed Three-Stranded Coiled Coils.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JH0
 
 | | Human Thrombin Hirugen Inhibitor complex | | Descriptor: | (2R)-2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}PROPENE-1-SULFONAMIDE, CALCIUM ION, HIRUDIN IIIA, ... | | Authors: | Senger, S, Chan, C, Convery, M.A, Hubbard, J.A, Young, R.J, Shah, G.P, Watson, N.S. | | Deposit date: | 2007-02-19 | | Release date: | 2007-05-08 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sulfonamide-Related Conformational Effects and Their Importance in Structure-Based Design.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2JVK
 
 | |
