4FRL
 
 | | Crystal Structure of BBBB+UDP+Gal at pH 8.0 with MPD as the cryoprotectant | | Descriptor: | Histo-blood group ABO system transferase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Johal, A.R, Alfaro, J.A, Blackler, R.J, Schuman, B, Borisova, S.N, Evans, S.V. | | Deposit date: | 2012-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | pH-induced conformational changes in human ABO(H) blood group glycosyltransferases confirm the importance of electrostatic interactions in the formation of the semi-closed state.
Glycobiology, 24, 2014
|
|
1ZJ2
 
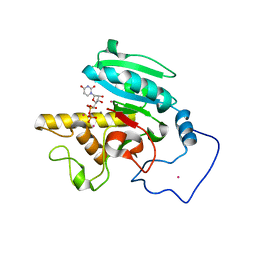 | | Crystal Structure of Human Galactosyltransferase (GTB) Complexed with H type I Trisaccharide | | Descriptor: | ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase), ... | | Authors: | Letts, J.A, Rose, N.L, Fang, Y.R, Barry, C.H, Borisova, S.N, Seto, N.O, Palcic, M.M, Evans, S.V. | | Deposit date: | 2005-04-27 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Differential Recognition of the Type I and II H Antigen Acceptors by the Human ABO(H) Blood Group A and B Glycosyltransferases.
J.Biol.Chem., 281, 2006
|
|
4F28
 
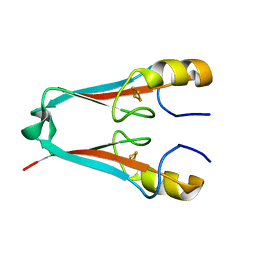 | | The Crystal Structure of a Human MitoNEET mutant with Met 62 Replaced by a Gly | | Descriptor: | CDGSH iron-sulfur domain-containing protein 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Baxter, E.L, Zuris, J.A, Wang, C, Axelrod, H.L, Cohen, A.E, Paddock, M.L, Nechushtai, R, Onuchic, J.N, Jennings, P.A. | | Deposit date: | 2012-05-07 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Allosteric control in a metalloprotein dramatically alters function.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1DT1
 
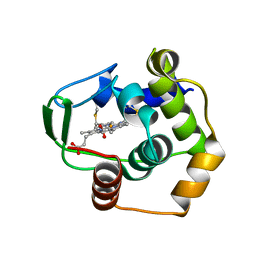 | | THERMUS THERMOPHILUS CYTOCHROME C552 SYNTHESIZED BY ESCHERICHIA COLI | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Fee, J.A, Chen, Y, Hill, M.J, Gomez-Moran, E, Loehr, T, Ai, J, Thony-Meyer, L, Williams, P.A, Stura, E, Sridhar, V, McRee, D.E. | | Deposit date: | 2000-01-10 | | Release date: | 2000-02-18 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integrity of thermus thermophilus cytochrome c552 synthesized by Escherichia coli cells expressing the host-specific cytochrome c maturation genes, ccmABCDEFGH: biochemical, spectral, and structural characterization of the recombinant protein.
Protein Sci., 9, 2000
|
|
2W9X
 
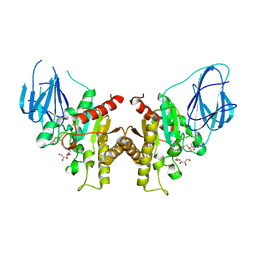 | | The active site of a carbohydrate esterase displays divergent catalytic and non-catalytic binding functions | | Descriptor: | GLYCEROL, PUTATIVE ACETYL XYLAN ESTERASE | | Authors: | Montanier, C, Money, V.A, Pires, V, Flint, J.E, Benedita, P.A, Goyal, A, Prates, J.A, Izumi, A, Stalbrand, H, Morland, C, Cartmell, A, Kolenova, K, Topakas, E, Dobson, E, Bolam, D.N, Davies, G.J, Fontes, C.M, Gilbert, H.J. | | Deposit date: | 2009-01-29 | | Release date: | 2009-03-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
2VW1
 
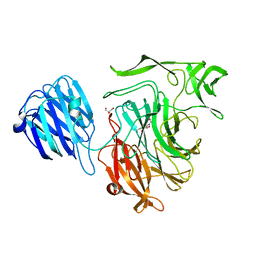 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
4FKI
 
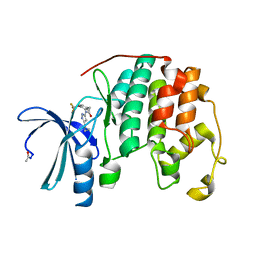 | |
4FKT
 
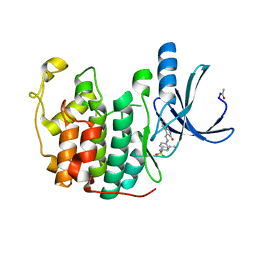 | |
2W2B
 
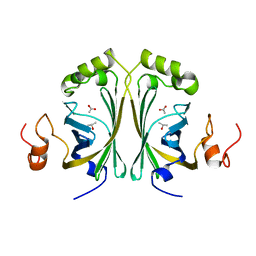 | | Crystal Structure of single point mutant Tyr20Phe p-coumaric Acid Decarboxylase from Lactobacillus plantarum: structural insights into the active site and decarboxylation catalytic mechanism | | Descriptor: | ACETATE ION, ISOPROPYL ALCOHOL, P-COUMARIC ACID DECARBOXYLASE | | Authors: | Rodriguez, H, Angulo, I, de las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-10-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
1XCS
 
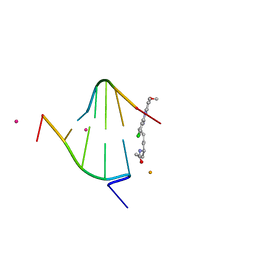 | | structure of oligonucleotide/drug complex | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-[(5-(ACETYLAMINO)-6-{[(1S,4R)-8-AMINO-4-[((2R)-6-AMINO-2-{2-[(1S)-5-AMINO-1-FORMYLPENTYL]HYDRAZINO}HEXANOYL)AMINO]-1-(4-AMINOBUTYL)-2,3-DIOXOOCTYL]AMINO}-6-OXOHEXYL)AMINO]-6-CHLORO-2-METHOXYACRIDINIUM, BARIUM ION, ... | | Authors: | Valls, N, Steiner, R.A, Wright, G, Murshudov, G.N, Subirana, J.A. | | Deposit date: | 2004-09-03 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Variable role of ions in two drug intercalation complexes of DNA
J.Biol.Inorg.Chem., 10, 2005
|
|
1OQV
 
 | |
2W06
 
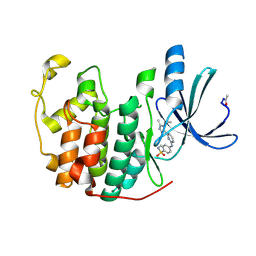 | | Structure of CDK2 in complex with an imidazolyl pyrimidine, compound 5c | | Descriptor: | 4-{[4-(1-CYCLOPROPYL-2-METHYL-1H-IMIDAZOL-5-YL)PYRIMIDIN-2-YL]AMINO}-N-METHYLBENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Anderson, M, Andrews, D.M, Barker, A.J, Brassington, C.A, Byth, K.F, Culshaw, J.D, Finlay, M.R.V, Fisher, E, Mcmiken, H.H.J, Green, C.P, Heaton, D.W, Nash, I.A, Newcombe, N.J, Oakes, S.E, Roberts, A, Stanway, J.J, Thomas, A.P, Tucker, J.A, Weir, H.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Imidazoles: Sar and Development of a Potent Class of Cyclin-Dependent Kinase Inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1OZU
 
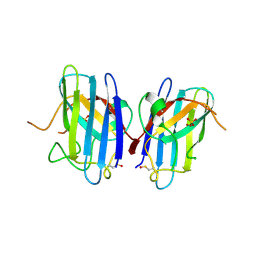 | | Crystal Structure of Familial ALS Mutant S134N of human Cu,Zn Superoxide Dismutase (CuZnSOD) to 1.3A resolution | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Elam, J.S, Taylor, A.B, Strange, R, Antonyuk, S, Doucette, P.A, Rodriguez, J.A, Hasnain, S.S, Hayward, L.J, Valentine, J.S, Yeates, T.O, Hart, P.J. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Amyloid-like Filaments and Water-filled Nanotubes Formed by SOD1 Mutant Proteins Linked to Familial ALS
Nat.Struct.Biol., 10, 2003
|
|
1CNB
 
 | |
1XBV
 
 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, MAGNESIUM ION, RIBULOSE-5-PHOSPHATE | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1X7G
 
 | | Actinorhodin Polyketide Ketoreductase, act KR, with NADP bound | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketoacyl reductase | | Authors: | Korman, T.P, Hill, J.A, Vu, T.N. | | Deposit date: | 2004-08-13 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity
Biochemistry, 43, 2004
|
|
1X7H
 
 | | Actinorhodin Polyketide Ketoreductase, with NADPH bound | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketoacyl reductase | | Authors: | Korman, T.P, Hill, J.A, Vu, T.N. | | Deposit date: | 2004-08-13 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity
Biochemistry, 43, 2004
|
|
2Y2W
 
 | | Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. | | Descriptor: | ARABINOFURANOSIDASE | | Authors: | Lagaert, S, Schoepe, J, Delcour, J.A, Lavigne, R, Strelkov, S.V, Courtin, C.M, Mikkelsen, N.E, Sandgren, M, Volckaert, G. | | Deposit date: | 2010-12-16 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elucidation of the Substrate Specificity and Protein Structure of Abfb, a Family 51 Alpha-L- Arabinofuranosidase from Bifidobacterium Longum.
To be Published
|
|
1X9A
 
 | | Solution NMR Structure of Protein Tm0979 from Thermotoga maritima. Ontario Center for Structural Proteomics Target TM0979_1_87; Northeast Structural Genomics Consortium Target VT98. | | Descriptor: | hypothetical protein TM0979 | | Authors: | Gaspar, J.A, Liu, C, Vassall, K.A, Stathopulos, P.B, Meglei, G, Stephen, R, Pineda-Lucena, A, Wu, B, Yee, A, Arrowsmith, C.H, Meiering, E.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-08-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel member of the YchN-like fold: solution structure of the hypothetical protein Tm0979 from Thermotoga maritima.
Protein Sci., 14, 2005
|
|
2XLJ
 
 | | Crystal structure of the Csy4-crRNA complex, hexagonal form | | Descriptor: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | Authors: | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
2XMA
 
 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE RIGHT END DNA COMPLEX | | Descriptor: | DRA2 TRANSPOSASE RIGHT END RECOGNITION SITE, MAGNESIUM ION, TRANSPOSASE | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
1CVE
 
 | |
1Q6L
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
2XLK
 
 | | Crystal structure of the Csy4-crRNA complex, orthorhombic form | | Descriptor: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | Authors: | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | Deposit date: | 2010-07-21 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
1XE3
 
 | | Crystal Structure of purine nucleoside phosphorylase DeoD from Bacillus anthracis | | Descriptor: | CHLORIDE ION, purine nucleoside phosphorylase | | Authors: | Grenha, R, Levdikov, V.M, Fogg, M, Blagova, E.V, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-09-09 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of purine nucleoside phosphorylase (DeoD) from Bacillus anthracis.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
