6EAX
 
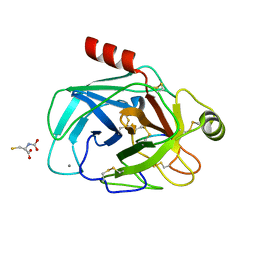 | | Crystallographic structure of the cyclic hexapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21 | | Descriptor: | CALCIUM ION, CYS-THR-LYS-SER-ILE-CYS, Cationic trypsin, ... | | Authors: | Fernandes, J.C, Valadares, N.F, Freitas, S.M, Barbosa, J.A.R.G. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.189 Å) | | Cite: | Crystallographic structure of the cyclic hexapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21
To Be Published
|
|
5G21
 
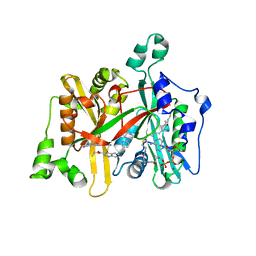 | | Leishmania major N-myristoyltransferase in complex with a quinoline inhibitor (compound 26). | | Descriptor: | ETHYL 4-[(2-CYANOETHYL)SULFANYL]-6-{[6-(PIPERAZIN-1-YL), GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
5G1Z
 
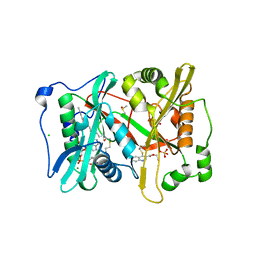 | | Plasmodium vivax N-myristoyltransferase in complex with a quinoline inhibitor (compound 1) | | Descriptor: | 2-oxopentadecyl-CoA, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Goncalves, V, Brannigan, J.A, Laporte, A, Bell, A.S, Roberts, S.M, Wilkinson, A.J, Leatherbarrow, R.J, Tate, E.W. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided optimization of quinoline inhibitors of Plasmodium N-myristoyltransferase.
Medchemcomm, 8, 2017
|
|
5BR1
 
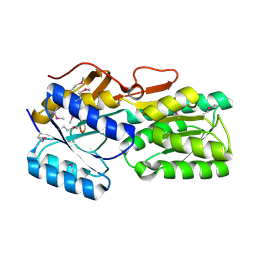 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM AGROBACTERIUM VITIS S4 (Avi_5305, TARGET EFI-511224) WITH BOUND ALPHA-D-GALACTOSAMINE | | Descriptor: | 2-amino-2-deoxy-alpha-D-galactopyranose, ABC transporter, binding protein | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of an ABC transporter solute-binding protein specific for the amino sugars glucosamine and galactosamine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5ANV
 
 | | MTH1 in complex with compound 15 | | Descriptor: | 4-(4-CHLORO-2-FLUORO-ANILINO)-6,7-DIMETHOXY-N-METHYL-QUINOLINE-3-CARBOXAMIDE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
5BT4
 
 | | Crystal structure of BRD4 first bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Bromodomain-containing protein 4 | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of BRD4 first bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
6E9D
 
 | | Sub-2 Angstrom Ewald Curvature-Corrected Single-Particle Cryo-EM Reconstruction of AAV-2 L336C | | Descriptor: | Capsid protein VP1 | | Authors: | Tan, Y.Z, Aiyer, S, Mietzsch, M, Hull, J.A, McKenna, R, Baker, T.S, Agbandje-McKenna, M, Lyumkis, D. | | Deposit date: | 2018-07-31 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (1.86 Å) | | Cite: | Sub-2 angstrom Ewald curvature corrected structure of an AAV2 capsid variant.
Nat Commun, 9, 2018
|
|
4CMP
 
 | | Crystal structure of S. pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation.
Science, 343, 2014
|
|
6ELR
 
 | | Human jak1 kinase domain in complex with compound 7 | | Descriptor: | SODIUM ION, Tyrosine-Protein Kinase JAK1, [3-[[5-methyl-2-[[3-(4-methylpiperazin-1-yl)-5-methylsulfonyl-phenyl]amino]pyrimidin-4-yl]amino]phenyl]methanol | | Authors: | Read, J.A. | | Deposit date: | 2017-09-29 | | Release date: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human jak1 kinase domain in complex with compound 7
To Be Published
|
|
4COS
 
 | | Crystal structure of the PHD-Bromo-PWWP cassette of human PRKCBP1 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, PROTEIN KINASE C-BINDING PROTEIN 1, ZINC ION | | Authors: | Krojer, T, Savitsky, P, Newman, J.A, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Filippakopoulos, P. | | Deposit date: | 2014-01-30 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Multivalent Histone and DNA Engagement by a PHD/BRD/PWWP Triple Reader Cassette Recruits ZMYND8 to K14ac-Rich Chromatin.
Cell Rep, 17, 2016
|
|
5G6T
 
 | | Crystal structure of Zn-containing NagZ H174A mutant from Pseudomonas aeruginosa | | Descriptor: | BETA-HEXOSAMINIDASE, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-07-15 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
4D0Y
 
 | |
6E9X
 
 | | DHF91 filament | | Descriptor: | DHF91 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6EAU
 
 | | Crystallographic structure of the octapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21. | | Descriptor: | CALCIUM ION, CYS-THR-LYS-SER-ILE-PRO-PRO-CYS, Cationic trypsin, ... | | Authors: | Fernandes, J.C, Valadares, N.F, Freitas, S.M, Barbosa, J.A.R.G. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystallographic structure of the octapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21.
To Be Published
|
|
6E6K
 
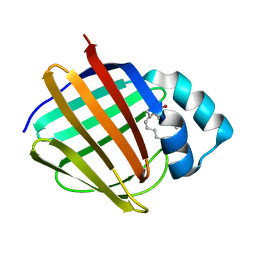 | | Crystal structure of human cellular retinol-binding protein 4 in complex with abnormal-cannabidiol (abn-CBD) | | Descriptor: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinoid-binding protein 7 | | Authors: | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6EEX
 
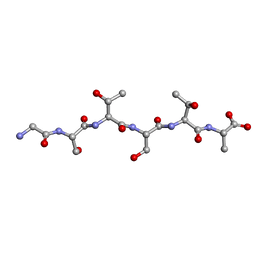 | | L-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | L-GSTSTA from ice nucleaction protein, inaZ | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-15 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
6EIU
 
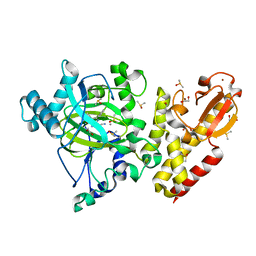 | | Crystal structure of KDM5B in complex with KDOPZ29a | | Descriptor: | 1,2-ETHANEDIOL, 8-oxidanyl-6-phenyl-7-propan-2-yl-imidazo[1,2-b]pyridazine-3-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Srikannathasan, V, Newman, J.A, Szykowska, A, Wright, M, Ruda, G.F, Vazquez-Rodriguez, S.A, Kupinska, K, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Huber, K, von Delft, F. | | Deposit date: | 2017-09-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of KDM5B in complex with KDOPZ29a.
to be published
|
|
4CIC
 
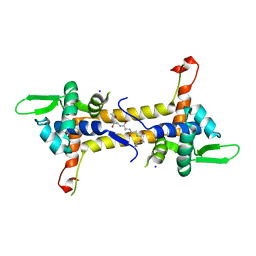 | | T. potens IscR | | Descriptor: | HEXA-ALANINE PEPTIDE, SODIUM ION, TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Santos, J.A, Macedo-Ribeiro, S, Pereira, P.J.B. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Unique Regulation of Iron-Sulfur Cluster Biogenesis in a Gram-Positive Bacterium.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CVD
 
 | |
5G3R
 
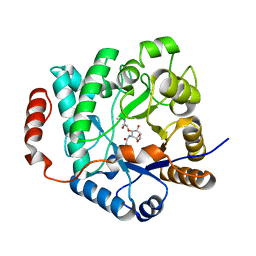 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine and L-Ala-1,6-anhydroMurNAc | | Descriptor: | 2-[[(2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-oxidanyl-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoyl]amino]propanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase, ... | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-04-30 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5BOP
 
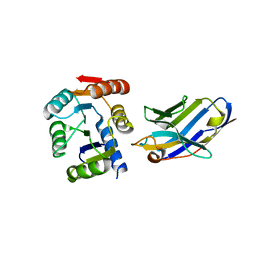 | | Crystal structure of the artificial nanobody octarellinV.1 complex | | Descriptor: | Nanobody, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Pardon, E, Steyaert, J, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-27 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
4CVO
 
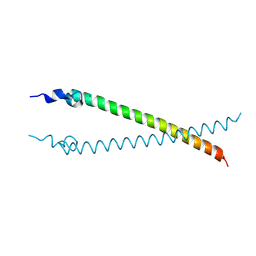 | | Crystal structure of the N-terminal colied-coil domain of human DNA excision repair protein ERCC-6 | | Descriptor: | DNA EXCISION REPAIR PROTEIN ERCC-6, MAGNESIUM ION | | Authors: | Newman, J.A, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2014-03-28 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the N-Terminal Colied-Coil Domain of Human DNA Excision Repair Protein Ercc-6
To be Published
|
|
5BQM
 
 | | Crystal structure of SXN101959, a Clostridium botulinum neurotoxin type D derivative and targeted secretion inhibitor | | Descriptor: | Botulinum neurotoxin type D, Somatoliberin,Botulinum neurotoxin type D, ZINC ION | | Authors: | Masuyer, G, Davies, J.R, Moore, K, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of Clostridium botulinum neurotoxin type D as a platform for the development of targeted secretion inhibitors.
Sci Rep, 5, 2015
|
|
6EAT
 
 | | Crystallographic structure of the cyclic nonapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21. | | Descriptor: | 9MER-PEPTIDE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Fernandes, J.C, Valadares, N.F, Freitas, S.M, Barbosa, J.A.R.G. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Crystallographic structure of a complex between trypsin and a nonapeptide derived from a Bowman-Birk inhibitor found in Vigna unguiculata seeds.
Arch. Biochem. Biophys., 665, 2019
|
|
4DEX
 
 | |
