6S9V
 
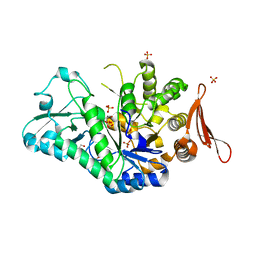 | | Crystal structure of sucrose 6F-phosphate phosphorylase from Thermoanaerobacter thermosaccharolyticum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Capra, N, Franceus, J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Comparison of a Promiscuous and a Highly Specific Sucrose 6 F -Phosphate Phosphorylase.
Int J Mol Sci, 20, 2019
|
|
4XXX
 
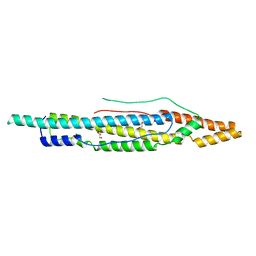 | | Structure of PE-PPE domains of ESX-1 secreted protein EspB, C2221 | | Descriptor: | CHLORIDE ION, ESX-1 secretion-associated protein EspB, GLYCEROL | | Authors: | Piton, J, Pojer, F, Korotkov, K.V. | | Deposit date: | 2015-01-30 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of EspB, a secreted substrate of the ESX-1 secretion system of Mycobacterium tuberculosis.
J.Struct.Biol., 191, 2015
|
|
8U95
 
 | | The structure of myosin heavy chain from Drosophila melanogaster flight muscle thick filaments | | Descriptor: | Myosin heavy chain, isoform U | | Authors: | Abbasi Yeganeh, F, Rastegarpouyani, H, Li, J, Taylor, K.A. | | Deposit date: | 2023-09-18 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the Drosophila melanogaster Flight Muscle Myosin Filament at 4.7 angstrom Resolution Reveals New Details of Non-Myosin Proteins.
Int J Mol Sci, 24, 2023
|
|
6SAZ
 
 | | Cleaved human fetuin-b in complex with crayfish astacin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Astacin, ... | | Authors: | Gomis-Ruth, F.X, Guevara, T, Cuppari, A, Korschgen, H, Schmitz, C, Kuske, M, Yiallouros, I, Floehr, J, Jahnen-Dechent, W, Stocker, W. | | Deposit date: | 2019-07-18 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C-terminal region of human plasma fetuin-B is dispensable for the raised-elephant-trunk mechanism of inhibition of astacin metallopeptidases.
Sci Rep, 9, 2019
|
|
4W4T
 
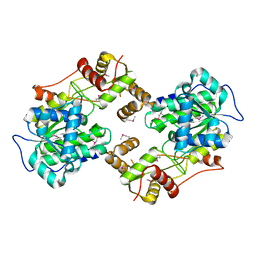 | | The crystal structure of the terminal R domain from the myxalamid PKS-NRPS biosynthetic pathway | | Descriptor: | ACETATE ION, MxaA | | Authors: | Tsai, S.C, Keasling, J.D, Luo, R, Barajas, J.F, Phelan, R.M, Schaub, A.J, Kliewer, J. | | Deposit date: | 2014-08-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Comprehensive Structural and Biochemical Analysis of the Terminal Myxalamid Reductase Domain for the Engineered Production of Primary Alcohols.
Chem.Biol., 22, 2015
|
|
4HJW
 
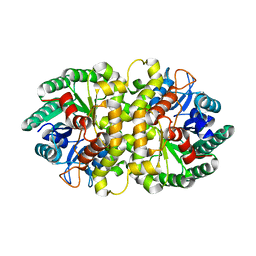 | | Crystal structure of Metarhizium anisopliae IDCase in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Zhu, J, Ding, J. | | Deposit date: | 2012-10-14 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
6POL
 
 | | Crystal structure of the human NELL1 EGF1-3-Robo3 FN1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, J, Pak, J.S, Ozkan, E. | | Deposit date: | 2019-07-04 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NELL2-Robo3 complex structure reveals mechanisms of receptor activation for axon guidance.
Nat Commun, 11, 2020
|
|
3FHD
 
 | | Crystal structure of the Shutoff and Exonuclease Protein from Kaposis Sarcoma Associated Herpesvirus | | Descriptor: | MAGNESIUM ION, ORF 37, SULFATE ION | | Authors: | Dahlroth, S.L, Gurmu, D, Schmitzberger, F, Haas, J, Erlandsen, H, Nordlund, P. | | Deposit date: | 2008-12-09 | | Release date: | 2009-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the shutoff and exonuclease protein from the oncogenic Kaposi's sarcoma-associated herpesvirus
Febs J., 276, 2009
|
|
6PI6
 
 | | The evolving story of AtzT, a periplasmic binding protein | | Descriptor: | 4-(ethylamino)-6-[(propan-2-yl)amino]-1,3,5-triazin-2-ol, Atrazine periplasmic binding protein, DIMETHYL SULFOXIDE | | Authors: | Peat, T.S, Newman, J, Scott, C, Esquirol, L, Dennis, M, Nebl, T. | | Deposit date: | 2019-06-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The evolving story of AtzT, a periplasmic binding protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4QF1
 
 | | Crystal structure of unliganded CH59UA, the inferred unmutated ancestor of the RV144 anti-HIV antibody lineage producing CH59 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CH59UA Fab fragment of heavy chain, CHLORIDE ION, ... | | Authors: | Wiehe, K, Easterhoff, D, Luo, K, Nicely, N.I, Bradley, T, Jaeger, F.H, Dennison, S.M, Zhang, R, Lloyd, K.E, Stolarchuk, C, Parks, R, Sutherland, L.L, Scearce, R.M, Morris, L, Kaewkungwal, J, Nitayaphan, S, Pitisuttithum, P, Rerks-Ngarm, S, Michael, N, Kim, J, Kelsoe, G, Montefiori, D.C, Tomaras, G, Bonsignori, M, Santra, S, Kepler, T.B, Alam, S.M, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2014-05-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antibody Light-Chain-Restricted Recognition of the Site of Immune Pressure in the RV144 HIV-1 Vaccine Trial Is Phylogenetically Conserved.
Immunity, 41, 2014
|
|
6PRF
 
 | | HIV-1 Protease multiple drug resistant clinical isolate mutant PR20 with GRL-14213A | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease, YTTRIUM ION | | Authors: | Kneller, D.W, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor combats highly drug resistant mutant PR20.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
6PKS
 
 | | MicroED structure of proteinase K from low-dose merged lamellae that were not pre-coated with platinum 2.16A resolution (LD) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.16 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
8UDG
 
 | | S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004 | | Descriptor: | Hemagglutinin, S1V2-72 heavy chain, S1V2-72 light chain | | Authors: | Finney, J, Kong, S, Walsh Jr, R.M, Harrison, S.C, Kelsoe, G. | | Deposit date: | 2023-09-28 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (4.98 Å) | | Cite: | Protective human antibodies against a conserved epitope in pre- and postfusion influenza hemagglutinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6PPK
 
 | | RbgA+45SRbgA complex | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Ortega, J. | | Deposit date: | 2019-07-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate.
Nucleic Acids Res., 47, 2019
|
|
4HZ2
 
 | | Crystal structure of glutathione s-transferase xaut_3756 (target efi-507152) from xanthobacter autotrophicus py2 | | Descriptor: | BENZOIC ACID, GLUTATHIONE, Glutathione S-transferase domain, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-11-14 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of glutathione s-transferase xaut_3756 from xanthobacter autotrophicus py2
To be Published
|
|
1U6B
 
 | | CRYSTAL STRUCTURE OF A SELF-SPLICING GROUP I INTRON WITH BOTH EXONS | | Descriptor: | 197-MER, 5'-R(*AP*AP*GP*CP*CP*AP*CP*AP*CP*AP*AP*AP*CP*CP*AP*GP*AP*CP*GP *GP*CP*C)-3', 5'-R(*CP*AP*(5MU))-3', ... | | Authors: | Adams, P.L, Stahley, M.R, Kosek, A.B, Wang, J, Strobel, S.A. | | Deposit date: | 2004-07-29 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Self-Splicing Group I Intron with Both Exons.
Nature, 430, 2004
|
|
3WUG
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
2NM2
 
 | |
3X13
 
 | | Crystal structure of HLA-B*0801.N80I | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2014-10-24 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The interaction of KIR3DL1*001 with HLA class I molecules is dependent upon molecular microarchitecture within the Bw4 epitope
J.Immunol., 194, 2015
|
|
8V1P
 
 | | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH UBF9092 | | Descriptor: | Glucose-induced degradation protein 4 homolog, N,N~2~-bis[(4-methoxyphenyl)methyl]glycinamide | | Authors: | Dong, C, Dong, A, Calabrese, M, Wang, F, Owen, D, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-21 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH UBF9092
To be published
|
|
5M8E
 
 | | Crystal structure of a GH43 arabonofuranosidase from Weissella sp. strain 142 | | Descriptor: | Alpha-N-arabinofuranosidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Linares-Pasten, J, Logan, D.T, Nordberg Karlsson, E. | | Deposit date: | 2016-10-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures and functional studies of two GH43 arabinofuranosidases from Weissella sp. strain 142 and Lactobacillus brevis.
FEBS J., 284, 2017
|
|
3ZVG
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 98 | | Descriptor: | 3C PROTEASE, N-(tert-butoxycarbonyl)-O-tert-butyl-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
4Y7R
 
 | | Crystal structure of WDR5 in complex with MYC MbIIIb peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MYC MbIIIb peptide, ... | | Authors: | Sun, Q, Phan, J, Olejniczak, E.T, Thomas, L.R, Fesik, S.W, Tansey, W.P. | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Interaction with WDR5 Promotes Target Gene Recognition and Tumorigenesis by MYC.
Mol.Cell, 58, 2015
|
|
3D6F
 
 | |
4YKC
 
 | |
