6YN5
 
 | | Inducible lysine decarboxylase LdcI decamer, pH 7.0 | | Descriptor: | Inducible lysine decarboxylase | | Authors: | Jessop, M, Felix, J, Desfosses, A, Effantin, G, Gutsche, I. | | Deposit date: | 2020-04-10 | | Release date: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YBS
 
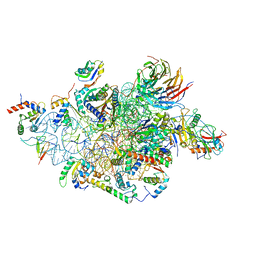 | | Structure of a human 48S translational initiation complex - head | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
7QNE
 
 | | Cryo-EM structure of human full-length synaptic alpha1beta3gamma2 GABA(A)R in complex with Ro15-4513 and megabody Mb38 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QND
 
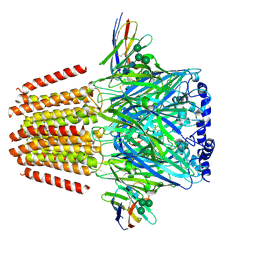 | | Cryo-EM structure of human full-length extrasynaptic beta3delta GABA(A)R in complex with THIP (gaboxadol), histamine and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,5,6,7-tetrahydro-[1,2]oxazolo[5,4-c]pyridin-3-one, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QNB
 
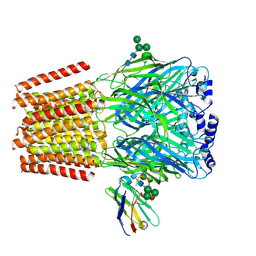 | | Cryo-EM structure of human full-length beta3gamma2 GABA(A)R in complex with GABA and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QNA
 
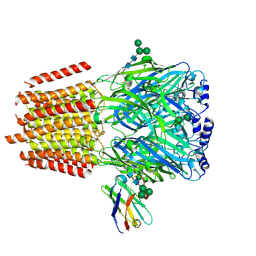 | | Cryo-EM structure of human full-length alpha4beta3gamma2 GABA(A)R in complex with GABA and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QN8
 
 | | Cryo-EM structure of human full-length beta3delta GABA(A)R in complex with histamine and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit beta-3, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QN6
 
 | | Cryo-EM structure of human full-length beta3delta GABA(A)R in complex with nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
7QN9
 
 | | Cryo-EM structure of human full-length extrasynaptic alpha4beta3delta GABA(A)R in complex with GABA, histamine and nanobody Nb25 in a pre-open/closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DECANE, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
6Y10
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant Q126H | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
6YN6
 
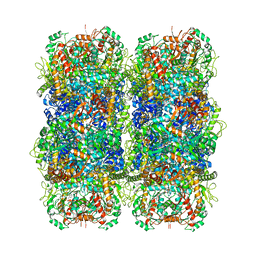 | | Inducible lysine decarboxylase LdcI stacks, pH 5.7 | | Descriptor: | Inducible lysine decarboxylase | | Authors: | Felix, J, Jessop, M, Desfosses, A, Effantin, G, Gutsche, I. | | Deposit date: | 2020-04-10 | | Release date: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4N1G
 
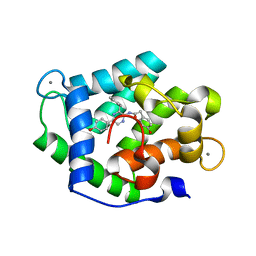 | | Crystal Structure of Ca(2+)- discharged F88Y obelin mutant from Obelia longissima at 1.50 Angstrom resolution | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Natashin, P.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-10-04 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the F88Y obelin mutant before and after bioluminescence provide molecular insight into spectral tuning among hydromedusan photoproteins
Febs J., 281, 2014
|
|
5CKY
 
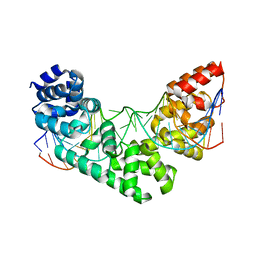 | | Crystal Structure of the MTERF1 R162A substitution bound to the termination sequence. | | Descriptor: | 5' -D (*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3', 5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3', Transcription termination factor 1, ... | | Authors: | Byrnes, J, Hauser, K, Norona, L, Mejia, E, Simmerling, C, Garcia-Diaz, M. | | Deposit date: | 2015-07-15 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Base Flipping by MTERF1 Can Accommodate Multiple Conformations and Occurs in a Stepwise Fashion.
J.Mol.Biol., 428, 2016
|
|
6Y6S
 
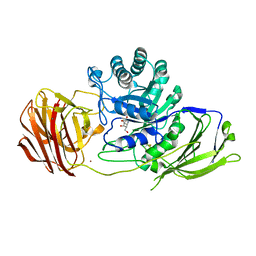 | | Mouse Galactocerebrosidase complexed with galacto-noeurostegine GNS at pH 6.8 | | Descriptor: | (1~{R},2~{S},3~{S},4~{R},5~{R})-4-(hydroxymethyl)-8-azabicyclo[3.2.1]octane-1,2,3-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Deane, J.E, McLoughlin, J. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Bicyclic Form of galacto -Noeurostegine Is a Potent Inhibitor of beta-Galactocerebrosidase.
Acs Med.Chem.Lett., 12, 2021
|
|
5KHL
 
 | |
1PBE
 
 | |
7QB1
 
 | | PPARg in complex with inhibitor | | Descriptor: | 3-[(3,4-dichlorophenyl)methyl]-4-oxidanylidene-6-phenoxy-phthalazine-1-carboxylic acid, Peroxisome proliferator-activated receptor gamma, SULFATE ION | | Authors: | Petersen, J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery by Virtual Screening of an Inhibitor of CDK5-Mediated PPAR gamma Phosphorylation.
Acs Med.Chem.Lett., 13, 2022
|
|
6YHH
 
 | | X-ray Structure of Flavobacterium johnsoniae chitobiase (FjGH20) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-N-acetylglucosaminidase-like protein Glycoside hydrolase family 20, GLYCEROL | | Authors: | Mazurkewich, S, Helland, R, MacKenzie, A, Eijsink, V.G.H, Pope, P.B, Branden, G, Larsbrink, J. | | Deposit date: | 2020-03-30 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights of the enzymes from the chitin utilization locus of Flavobacterium johnsoniae.
Sci Rep, 10, 2020
|
|
6YHV
 
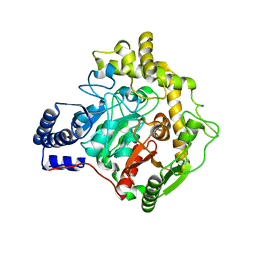 | | Structural insights into Pseudomonas aeruginosa Type six secretion system exported effector 8: unliganded Tse8 | | Descriptor: | COPPER (II) ION, Tse8 | | Authors: | Sainz-Polo, M.A, Capuni, R, Pretre, G, Gonzalez-Magana, A, Lucas, M, Altuna, J, Montanchez, I, Fucini, P, Albesa-Jove, D. | | Deposit date: | 2020-03-31 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Structural insights into Pseudomonas aeruginosaType six secretion system exported effector 8.
J.Struct.Biol., 212, 2020
|
|
7QSM
 
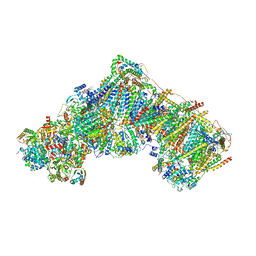 | | Bovine complex I in lipid nanodisc, Deactive-ligand (composite) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Chung, I, Bridges, H.R, Hirst, J. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-25 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSL
 
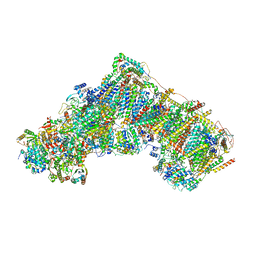 | | Bovine complex I in lipid nanodisc, Active-apo | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Chung, I, Bridges, H.R, Hirst, J. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-25 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QQC
 
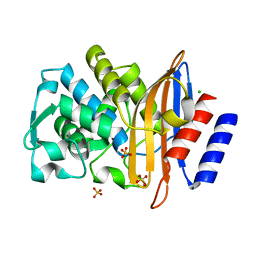 | | Structure of CTX-M-15 K73A mutant | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 13, 2022
|
|
7ZUV
 
 | | Crystal structure of Chlamydomonas reinhardtii chloroplastic sedoheptulose-1,7-bisphosphatase in reducing conditions | | Descriptor: | FBPase domain-containing protein, SULFATE ION | | Authors: | Le Moigne, T, Robert, G.Q, Lemaire, S.D, Henri, J. | | Deposit date: | 2022-05-13 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Characterization of chloroplast ribulose-5-phosphate-3-epimerase from the microalga Chlamydomonas reinhardtii.
Plant Physiol., 194, 2024
|
|
7QSN
 
 | | Bovine complex I in lipid nanodisc, Deactive-apo | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Chung, I, Bridges, H.R, Hirst, J. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-25 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
6JAU
 
 | | The complex structure of Pseudomonas aeruginosa MucA/MucB. | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Li, T, He, L.H, Li, C.C, Liu, L, Peng, C.T, Shen, Y.L, Qin, X.F, Xiao, Q.J, Zhu, Y.B, Song, Y.J, Zhao, N.l, Zhao, C, Yang, J, Mu, X.Y, Huang, Q, Bao, R. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Molecular basis of the lipid-induced MucA-MucB dissociation in Pseudomonas aeruginosa.
Commun Biol, 3, 2020
|
|
