4E4S
 
 | | Crystal structure of Pika GITRL | | Descriptor: | GLYCEROL, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 18 | | Authors: | Kumar, P.R, Bhosle, R, Bonanno, J, Chowdhury, S, Gizzi, A, Glen, S, Hillerich, B, Hammonds, J, Seidel, R, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of GITRL from Ochotona princeps
to be published
|
|
7T6I
 
 | | Crystal structure of HLA-DP1 in complex with pp65 peptide in reverse orientation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Lim, J.J, Reid, H, Rossjohn, J. | | Deposit date: | 2021-12-13 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human T cells recognize HLA-DP-bound peptides in two orientations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GYD
 
 | | Structure of Schistosoma japonicum Glutathione S-transferase bound with the ligand complex of 16 | | Descriptor: | (2R)-2-[[2-(5-chloranylthiophen-2-yl)-4-oxidanylidene-6-[2-(1H-1,2,3,4-tetrazol-5-yl)phenyl]quinazolin-3-yl]methyl]-3-(4-chlorophenyl)propanoic acid, ETHANOL, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Wen, X, Jin, R, Hu, H, Zhu, J, Song, W, Lu, X. | | Deposit date: | 2022-09-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery, SAR Study of GST Inhibitors from a Novel Quinazolin-4(1 H )-one Focused DNA-Encoded Library.
J.Med.Chem., 66, 2023
|
|
4E5T
 
 | | Crystal structure of a putative Mandelate racemase/Muconate lactonizing enzyme (Target PSI-200750) from Labrenzia alexandrii DFL-11 | | Descriptor: | MAGNESIUM ION, Mandelate racemase / muconate lactonizing enzyme, C-terminal domain protein | | Authors: | Kumar, P.R, Bonanno, J, Chowdhury, S, Foti, R, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Matikainen, B, Seidel, R, Toro, R, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-14 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a putative MR/ML enzyme from Labrenzia alexandrii DFL-11
to be published
|
|
4E69
 
 | | Crystal structure of a sugar kinase (target EFI-502132) from Oceanicola granulosus, unliganded structure | | Descriptor: | 1,2-ETHANEDIOL, 2-dehydro-3-deoxygluconokinase, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a sugar kinase (target EFI-502132) from Oceanicola granulosus, unliganded structure
To be Published
|
|
8GRJ
 
 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kojima, K, Tsugawa, W, Okuda-Shimazaki, J, Kerrigan, J.A, Sode, K. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone
To Be Published
|
|
4J30
 
 | | Structure of the effector - immunity system Tae4 / Tai4 from Salmonella typhimurium, selenomethionine variant | | Descriptor: | 2-ETHOXYETHANOL, CITRATE ANION, Putative cytoplasmic protein, ... | | Authors: | Benz, J, Reinstein, J, Meinhart, A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-05-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tae4/Tai4 from Salmonella typhimurium.
Plos One, 8, 2013
|
|
7TCV
 
 | | VDAC K12E mutant | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Khan, F, Abramson, J. | | Deposit date: | 2021-12-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Dynamical control of the mitochondrial beta-barrel channel VDAC by electrostatic and mechanical coupling
To Be Published
|
|
7TAK
 
 | | Structure of a NAT transporter | | Descriptor: | GUANINE, Putative membrane protein PurT | | Authors: | Weng, J, Zhou, X, Ren, Z, Chen, K, Zhou, M. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.79828 Å) | | Cite: | Structure of a NAT transporter
To Be Published
|
|
7T6E
 
 | |
7T90
 
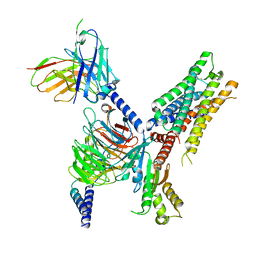 | | Cryo-EM structure of ACh-bound M2R-Go signaling complex in S2 state | | Descriptor: | ACETYLCHOLINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
7T8X
 
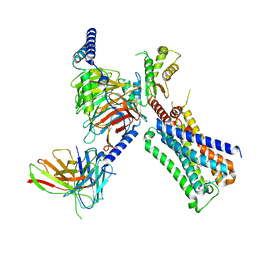 | | Cryo-EM structure of ACh-bound M2R-Go signaling complex in S1 state | | Descriptor: | ACETYLCHOLINE, Antibody fragment, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
8GQT
 
 | |
8GPR
 
 | |
7T96
 
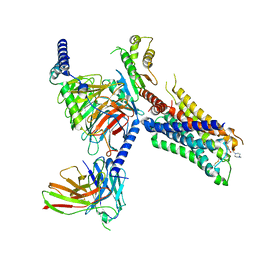 | | Cryo-EM structure of S2 state ACh-bound M2R-Go signaling complex with a PAM | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, ACETYLCHOLINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-18 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
1SH6
 
 | |
8GRW
 
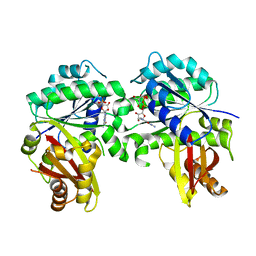 | | Spiroplasma melliferum FtsZ F224M bound to GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Chakraborty, J, Pananghat, G. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.40005422 Å) | | Cite: | Structural basis of kinetic polarity of FtsZ from cell wall less bacterium Spiroplasma
To Be Published
|
|
7T94
 
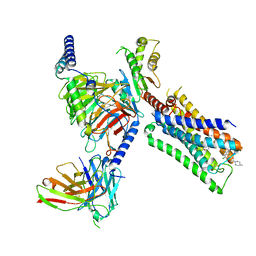 | | Cryo-EM structure of S1 state ACh-bound M2R-Go signaling complex with a PAM | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, ACETYLCHOLINE, Antibody fragment, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
8GX3
 
 | | The crystal structure of human Calpain-1 protease core in complex with 14c | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of human Calpain-1 protease core in complex with 14c
To Be Published
|
|
8ZF1
 
 | | Crystal structure of a Chemo Triplet Photoenzyme (CTPe) | | Descriptor: | N-(9-oxidanylidenethioxanthen-2-yl)ethanamide, Transcriptional regulator, PadR-like family | | Authors: | Chen, X, Qian, J.Y, Guo, J, Wu, Y.Z, Zhong, F.R. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chemogenetic Evolution of Diversified Photoenzymes for Enantioselective [2 + 2] Cycloadditions in Whole Cells.
J.Am.Chem.Soc., 2024
|
|
8GWD
 
 | | Crystal structure of AtHPPD-Y18734 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-[(1S)-1-phenylethyl]quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Crystal structure of AtHPPD-Y18734 complex
To Be Published
|
|
3FEA
 
 | |
8GWW
 
 | | Small-molecule Allosteric Regulation Mechanism of SHP2 | | Descriptor: | 2-[4-(aminomethyl)-4-methyl-piperidin-1-yl]-5-[2,3-bis(chloranyl)phenyl]-3-methyl-pyrrolo[2,1-f][1,2,4]triazin-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, Y, Zhu, J, Yu, K, Liu, B. | | Deposit date: | 2022-09-17 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Small-molecule Allosteric Regulation Mechanism of SHP2
To Be Published
|
|
8GEM
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with N-ethyl-N-({3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}methyl)-2-(1H-pyrazol-1-yl)ethanamine | | Descriptor: | N-ethyl-N-({3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}methyl)-2-(1H-pyrazol-1-yl)ethan-1-amine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GLI
 
 | | Crystal Structure of Human CD1b in Complex with Mycobacterial C85-GMM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-[(2R,3R)-3-hydroxy-20-{(1R,2S)-2-[(17R,18R)-17-methoxy-18-methyldotriacontyl]cyclopropyl}-2-pentadecylicosanoyl]-alpha-L-galactopyranose, Beta-2-microglobulin, ... | | Authors: | Balaji, G, Rossjohn, J, Shahine, A. | | Deposit date: | 2023-03-22 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CD1 lipidomes reveal lipid-binding motifs and size-based antigen-display mechanisms.
Cell, 186, 2023
|
|
