8FCG
 
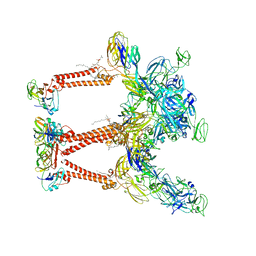 | | Cryo-EM structure of Chikungunya virus asymmetric unit | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Capsid protein, E1 glycoprotein, ... | | Authors: | Su, G.C, Chmielewsk, D, Kaelber, J, Pintilie, G, Chen, M, Jin, J, Auguste, A, Chiu, W. | | Deposit date: | 2022-12-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryogenic electron microscopy and tomography reveal imperfect icosahedral symmetry in alphaviruses.
Pnas Nexus, 3, 2024
|
|
5DMH
 
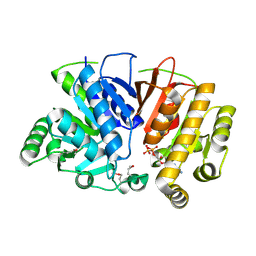 | | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Uncharacterized protein conserved in bacteria | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-08 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP.
To be published
|
|
6U3M
 
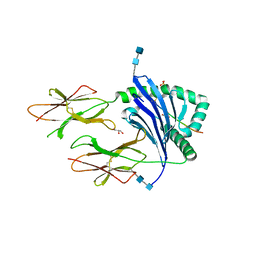 | | DQ2-P.fluor-alpha1a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha1a peptide, ... | | Authors: | Petersen, J, Rossjohn, J. | | Deposit date: | 2019-08-22 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | T cell receptor cross-reactivity between gliadin and bacterial peptides in celiac disease.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2D47
 
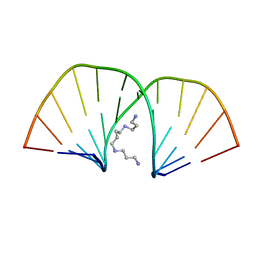 | | MOLECULAR STRUCTURE OF A COMPLETE TURN OF A-DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*GP*CP*GP*GP*GP*GP*G)-3'), SPERMINE | | Authors: | Verdaguer, N, Aymami, J, Fernandez-Forner, D, Fita, I, Coll, M, Huynh-Dinh, T, Igolen, J, Subirana, J.A. | | Deposit date: | 1991-10-02 | | Release date: | 1991-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of a complete turn of A-DNA.
J.Mol.Biol., 221, 1991
|
|
3DBC
 
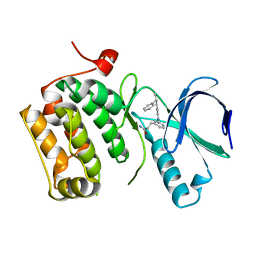 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 257 | | Descriptor: | 3-[3-(3-methyl-6-{[(1S)-1-phenylethyl]amino}-1H-pyrazolo[4,3-c]pyridin-1-yl)phenyl]propanamide, Polo-like kinase 1 | | Authors: | Elling, R.A, Zhu, J, Barr, K.J, Romanowski, M.J. | | Deposit date: | 2008-05-31 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Design and synthesis of 2-amino-pyrazolopyridines as Polo-like kinase 1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6HME
 
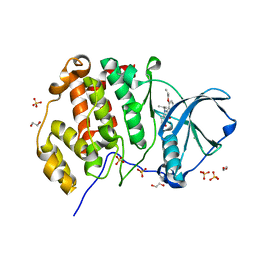 | | LOW-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA; CSNK2A1 gene product) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR THN27 | | Descriptor: | 1,2-ETHANEDIOL, 5-propan-2-yl-4-prop-2-enoxy-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
6ZU9
 
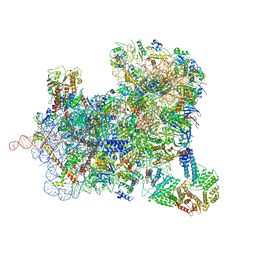 | | Structure of a yeast ABCE1-bound 48S initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
3MEU
 
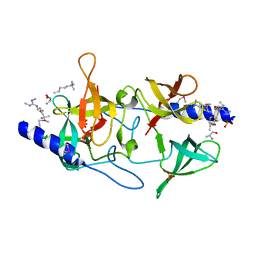 | | Crystal structure of SGF29 in complex with H3R2me2sK4me3 | | Descriptor: | Histone H3, SAGA-associated factor 29 homolog, SULFATE ION | | Authors: | Bian, C.B, Xu, C, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
6ZOQ
 
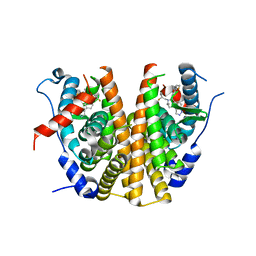 | | Oestrogen receptor ligand binding domain in complex with compound 16 | | Descriptor: | Estrogen receptor, ~{N}-[4-[(6~{S},8~{R})-7-[(1-fluoranylcyclopropyl)methyl]-8-methyl-2,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]-3-methoxy-phenyl]-1-(3-fluoranylpropyl)azetidin-3-amine | | Authors: | Breed, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
J.Med.Chem., 63, 2020
|
|
3MEV
 
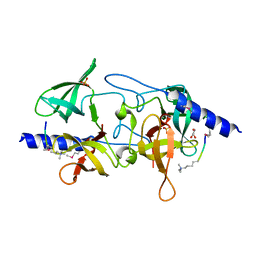 | | Crystal structure of SGF29 in complex with R2AK4me3 | | Descriptor: | GLYCEROL, Histone H3, SAGA-associated factor 29 homolog, ... | | Authors: | Bian, C.B, Xu, C, Lam, R, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
3DKF
 
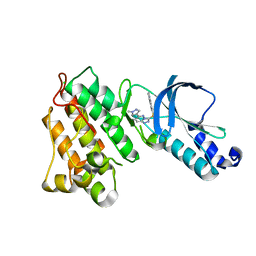 | | Structure of MET receptor tyrosine kinase in complex with inhibitor SGX-523 | | Descriptor: | 6-{[6-(1-methyl-1H-pyrazol-4-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}quinoline, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Hendle, J. | | Deposit date: | 2008-06-24 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SGX523 is an exquisitely selective, ATP-competitive inhibitor of the MET receptor tyrosine kinase with antitumor activity in vivo.
Mol.Cancer Ther., 8, 2009
|
|
3D94
 
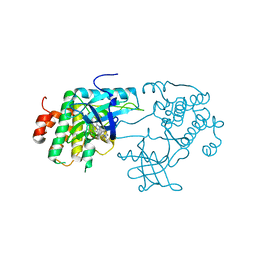 | | Crystal structure of the insulin-like growth factor-1 receptor kinase in complex with PQIP | | Descriptor: | 3-[cis-3-(4-methylpiperazin-1-yl)cyclobutyl]-1-(2-phenylquinolin-7-yl)imidazo[1,5-a]pyrazin-8-amine, CALCIUM ION, Insulin-like growth factor 1 receptor beta chain | | Authors: | Wu, J, Li, W, Miller, W.T, Hubbard, S.R. | | Deposit date: | 2008-05-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibition and activation-loop trans-phosphorylation of the IGF1 receptor
Embo J., 27, 2008
|
|
3MP1
 
 | | Complex structure of Sgf29 and trimethylated H3K4 | | Descriptor: | ACETATE ION, H3K4me3 peptide, Maltose-binding periplasmic protein,LINKER,SAGA-associated factor 29, ... | | Authors: | Li, J, Ruan, J, Wu, M, Xue, X, Zang, J. | | Deposit date: | 2010-04-24 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation
Embo J., 30, 2011
|
|
7A54
 
 | |
5DOI
 
 | | Crystal structure of Tetrahymena p45N and p19 | | Descriptor: | Telomerase associated protein p45, Telomerase-associated protein 19 | | Authors: | Wan, B, Tang, T, Wu, J, Lei, M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
6ZPR
 
 | | Solution structure of MLKL executioner domain in complex with a covalent inhibitor | | Descriptor: | 7-(2-methoxyethoxymethyl)-1,3-dimethyl-purine-2,6-dione, Mixed lineage kinase domain-like protein,Mixed lineage kinase domain-like protein | | Authors: | Ruebbelke, M, Bauer, M, Hamilton, J, Binder, F, Nar, H, Zeeb, M. | | Deposit date: | 2020-07-09 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-06 | | Method: | SOLUTION NMR | | Cite: | Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZZZ
 
 | |
7A0V
 
 | | Crystal structure of the 5-phosphatase domain of Synaptojanin1 in complex with a nanobody | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nanobody 13015, ... | | Authors: | Paesmans, J, Galicia, C, Martin, E, Versees, W. | | Deposit date: | 2020-08-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structure of substrate-bound Synaptojanin1 provides new insights in its mechanism and the effect of disease mutations.
Elife, 9, 2020
|
|
5DOK
 
 | | Crystal structure of Tetrahymena p45C | | Descriptor: | MAGNESIUM ION, Telomerase associated protein p45 | | Authors: | Wan, B, Tang, T, Wu, J, Lei, M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
3DBF
 
 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 562 | | Descriptor: | 4-({1-[3-(3-amino-3-oxopropyl)-5-chlorophenyl]-3-methyl-1H-pyrazolo[4,3-c]pyridin-6-yl}amino)-3-methoxy-N-(1-methylpipe ridin-4-yl)benzamide, Polo-like kinase | | Authors: | Elling, R.A, Zhu, J, Barr, K.J, Romanowski, M.J. | | Deposit date: | 2008-05-31 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 2-amino-pyrazolopyridines as Polo-like kinase 1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4FBY
 
 | | fs X-ray diffraction of Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Hellmich, J, Tran, R, Hattne, J, Laksmono, H, Gloeckner, C, Echols, N, Sierra, R.G, Sellberg, J, Lassalle-Kaiser, B, Gildea, R.J, Glatzel, P, Grosse-Kunstleve, R.W, Latimer, M.J, Mcqueen, T.A, Difiore, D, Fry, A.R, Messerschmidt, M.M, Miahnahri, A, Schafer, D.W, Seibert, M.M, Sokaras, D, Weng, T.-C, Zwart, P.H, White, W.E, Adams, P.D, Bogan, M.J, Boutet, S, Williams, G.J, Messinger, J, Sauter, N.K, Zouni, A, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (6.56 Å) | | Cite: | Room temperature femtosecond X-ray diffraction of photosystem II microcrystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MEA
 
 | | Crystal structure of the SGF29 in complex with H3K4me3 | | Descriptor: | Histone H3, SAGA-associated factor 29 homolog | | Authors: | Bian, C, Xu, C, Tempel, W, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
7A60
 
 | | Crystal structure of VIM-2 with hydrolyzed faropenem (ring-open form) | | Descriptor: | (5~{Z})-2-[1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-5-(4-oxidanylbutylidene)-2~{H}-1,3-thiazole-4-carboxylic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
7A61
 
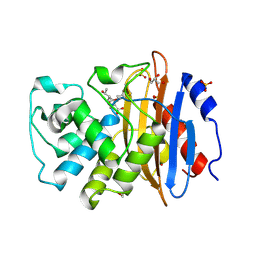 | | Crystal structure of KPC-2 with hydrolyzed faropenem (ring-open form) | | Descriptor: | (2~{R})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-5-butyl-2,3-dihydro-1,3-thiazole-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
7A63
 
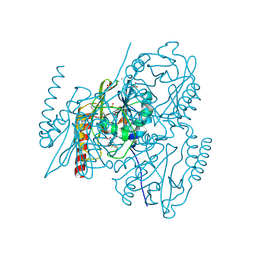 | | Crystal structure of L1 with hydrolyzed faropenem (imine, ring-closed form) | | Descriptor: | (2R,5S)-2-[(1S,2R)-1-carboxy-2-hydroxy-propyl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydrothiazole-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57000113 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
