3JZR
 
 | |
3VFV
 
 | | crystal structure of HLA B*3508 LPEP-P9Ala, peptide mutant P9-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P9A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
4CD4
 
 | | The structure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManIFG | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-1,4-BETA MANNANASE, PUTATIVE, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4FGB
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I apo form | | Descriptor: | Calcium/calmodulin-dependent protein kinase type 1 | | Authors: | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
3VFW
 
 | | crystal structure of HLA B*3508 LPEP-P10Ala, peptide mutant P10-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P10A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
4L9K
 
 | | X-ray study of human serum albumin complexed with camptothecin | | Descriptor: | (2S)-2-hydroxy-2-[8-(hydroxymethyl)-9-oxo-9,11-dihydroindolizino[1,2-b]quinolin-7-yl]butanoic acid, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-24 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4BU4
 
 | | RIBONUCLEASE T1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-14 | | Release date: | 1998-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
7YQ3
 
 | | human insulin receptor bound with A43 DNA aptamer and insulin | | Descriptor: | IR-A43 aptamer, Insulin A chain, Insulin, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
4C36
 
 | | PKA-S6K1 Chimera with compound 15e (CCT147581) bound | | Descriptor: | 4-[1-(cyclopropylmethyl)-1H-benzimidazol-2-yl]-1,2,5-oxadiazol-3-amine, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, ... | | Authors: | Couty, S, Westwood, I.M, Kalusa, A, Cano, C, Travers, J, Boxall, K, Chow, C.L, Burns, S, Schmitt, J, Pickard, L, Barillari, C, McAndrew, P.C, Clarke, P.A, Linardopoulos, S, Griffin, R.J, Aherne, G.W, Raynaud, F.I, Workman, P, Jones, K, van Montfort, R.L.M. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The discovery of potent ribosomal S6 kinase inhibitors by high-throughput screening and structure-guided drug design.
Oncotarget, 4, 2013
|
|
1T8M
 
 | | CRYSTAL STRUCTURE OF THE P1 HIS BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Czapinska, H, Helland, R, Otlewski, J, Smalas, A.O. | | Deposit date: | 2004-05-13 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of five bovine chymotrypsin complexes with P1 BPTI variants.
J.Mol.Biol., 344, 2004
|
|
7YQ4
 
 | | human insulin receptor bound with A62 DNA aptamer and insulin - locally refined | | Descriptor: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
4CA0
 
 | | Structural Basis for the microtubule binding of the human kinetochore Ska complex | | Descriptor: | SPINDLE AND KINETOCHORE-ASSOCIATED PROTEIN 1 | | Authors: | Abad, M, Medina, B, Santamaria, A, Zou, J, Plasberg-Hill, C, Madhumalar, A, Jayachandran, U, Redli, P.M, Rappsilber, J, Nigg, E.A, Jeyaprakash, A.A. | | Deposit date: | 2013-10-04 | | Release date: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (2.259 Å) | | Cite: | Structural Basis for Microtubule Recognition by the Human Kinetochore Ska Complex.
Nat.Commun., 5, 2014
|
|
1TAZ
 
 | | Catalytic Domain Of Human Phosphodiesterase 1B | | Descriptor: | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ZINC ION | | Authors: | Zhang, K.Y.J, Card, G.L, Suzuki, Y, Artis, D.R, Fong, D, Gillette, S, Hsieh, D, Neiman, J, West, B.L, Zhang, C, Milburn, M.V, Kim, S.-H, Schlessinger, J, Bollag, G. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Glutamine Switch Mechanism for Nucleotide Selectivity by Phosphodiesterases
Mol.Cell, 15, 2004
|
|
7YQ6
 
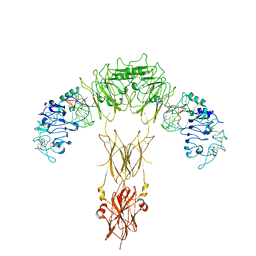 | | human insulin receptor bound with A62 DNA aptamer | | Descriptor: | IR-A62 aptamer, Isoform Short of Insulin receptor | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
7YQ5
 
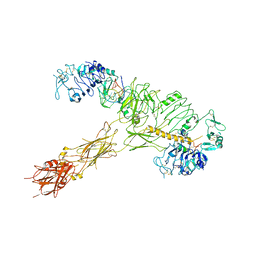 | | human insulin receptor bound with A62 DNA aptamer and insulin | | Descriptor: | IR-A62 aptamer, Insulin A chain, Insulin, ... | | Authors: | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
4CBI
 
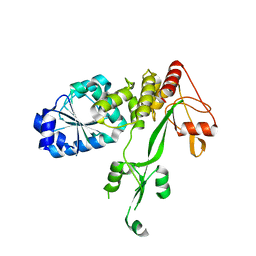 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
4BZ2
 
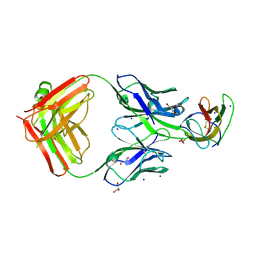 | | Structure of dengue virus EDIII in complex with Fab 2D73 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ENVELOPE PROTEIN, ... | | Authors: | Li, J, Watterson, D, Chang, C.W, Li, X.Q, Ericsson, D.J, Qiu, L.W, Cai, J.P, Chen, J, Fry, S.R, Cooper, M.A, Che, X.Y, Young, P.R, Kobe, B. | | Deposit date: | 2013-07-23 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of Dengue Virus Ediii in Complex with Fab 2D73
To be Published
|
|
3VFT
 
 | | crystal structure of HLA B*3508LPEP-P6Ala, peptide mutant P6-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P6A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
4CBH
 
 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
4GP3
 
 | | The crystal structure of human fascin 1 K358A mutant | | Descriptor: | BROMIDE ION, CHLORIDE ION, Fascin, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
2POE
 
 | | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660 | | Descriptor: | Cyclophilin-like protein, putative, FORMIC ACID | | Authors: | Wernimont, A.K, Lew, J, Hills, T, Hassanali, A, Lin, L, Wasney, G, Zhao, Y, Kozieradzki, I, Vedadi, M, Schapira, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-26 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Cryptosporidium parvum cyclophilin type peptidyl-prolyl cis-trans isomerase cgd2_1660.
To be Published
|
|
4GP0
 
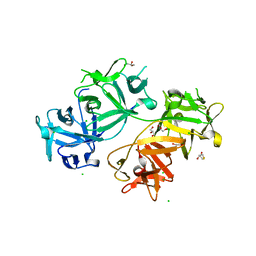 | | The crystal structure of human fascin 1 R149A K150A R151A mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
4CBM
 
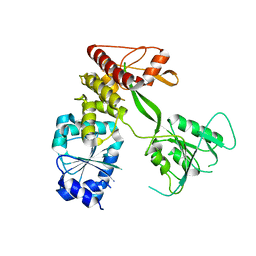 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
3VFP
 
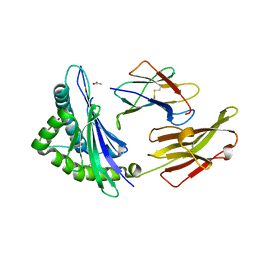 | | crystal structure of HLA B*3508 LPEP158G, HLA mutant Gly158 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, LPEP peptide from EBV, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
2Q3G
 
 | | Structure of the PDZ domain of human PDLIM7 bound to a C-terminal extension from human beta-tropomyosin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PDZ and LIM domain protein 7 | | Authors: | Gileadi, C, Papagrigoriou, E, Elkins, J, Burgess-Brown, N, Salah, E, Gileadi, O, Umeano, C, Bunkoczi, G, von Delft, F, Uppenberg, J, Pike, A.C.W, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms
Protein Sci., 19, 2010
|
|
