8TNH
 
 | |
2MUY
 
 | | The solution structure of the FtsH periplasmic N-domain | | Descriptor: | ATP-dependent zinc metalloprotease FtsH | | Authors: | Scharfenberg, F, Serek-Heuberger, J, Martin, J, Lupas, A.N, Coles, M. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Evolution of N-domains in AAA Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
8XQW
 
 | | Cryo-EM structure of the Ycf2-FtsHi motor complex from Chlamydomonas reinhardtii in AMPPNP bound state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ACP, ... | | Authors: | Liang, K, Zhan, X, Wu, J, Yan, Z. | | Deposit date: | 2024-01-10 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conservation and specialization of the Ycf2-FtsHi chloroplast protein import motor in green algae.
Cell, 2024
|
|
7DD3
 
 | | Cryo-EM structure of the pre-mRNA-loaded DEAH-box ATPase/helicase Prp2 in complex with Spp2 | | Descriptor: | PRP2 isoform 1, Pre-mRNA-splicing factor SPP2, pre-mRNA | | Authors: | Bai, R, Wan, R, Yan, C, Qi, J, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | Descriptor: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
3PCU
 
 | | Crystal structure of human retinoic X receptor alpha ligand-binding domain complexed with LX0278 and SRC1 peptide | | Descriptor: | 2-[(2S)-6-(2-methylbut-3-en-2-yl)-7-oxo-2,3-dihydro-7H-furo[3,2-g]chromen-2-yl]propan-2-yl acetate, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Zhang, H, Zhang, Y, Shen, H, Chen, J, Li, C, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-10-22 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (+)-Rutamarin as a Dual Inducer of Both GLUT4 Translocation and Expression Efficiently Ameliorates Glucose Homeostasis in Insulin-Resistant Mice.
Plos One, 7, 2012
|
|
8XI3
 
 | | Structure of mouse SCMC-14-3-3gama complex | | Descriptor: | 14-3-3 protein gamma, NACHT, LRR and PYD domains-containing protein 5, ... | | Authors: | Chi, P, Han, Z, Jiao, H, Li, J, Wang, X, Hu, H, Deng, D. | | Deposit date: | 2023-12-19 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of mouse SCMC-14-3-3gama complex
To Be Published
|
|
8WK2
 
 | | Crystal structure of AtHPPD-F419Y+Y13287 complex | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-26 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Crystal structure of AtHPPD-F419Y+Y13287 complex
To Be Published
|
|
8TNI
 
 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP gp120, ... | | Authors: | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex
To Be Published
|
|
4FBX
 
 | | Complex structure of human protein kinase CK2 catalytic subunit crystallized in the presence of a bisubstrate inhibitor | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha, bisubstrate inhibitor | | Authors: | Enkvist, E, Viht, K, Bischoff, N, Vahter, J, Saaver, S, Raidaru, G, Issinger, O.-G, Niefind, K, Uri, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A subnanomolar fluorescent probe for protein kinase CK2 interaction studies.
Org.Biomol.Chem., 10, 2012
|
|
8WL0
 
 | | Crystal structure of AtHPPD+Y19542 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 6-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-3-[(4-fluorophenyl)methyl]-5-methyl-1,2,3-benzotriazin-4-one, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-29 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of AtHPPD-+Y19542 complex
To Be Published
|
|
1VIT
 
 | | THROMBIN:HIRUDIN 51-65 COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA THROMBIN, ... | | Authors: | Vitali, J, Edwards, B.F.P. | | Deposit date: | 1996-01-31 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a bovine thrombin-hirudin51-65 complex determined by a combination of molecular replacement and graphics. Incorporation of known structural information in molecular replacement.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2IFV
 
 | |
8WJZ
 
 | | Crystal structure of AtHPPD-F419Y+Y13161 complex | | Descriptor: | 3-(2,6-dimethylphenyl)-1-methyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-26 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Crystal structure of AtHPPD-F419Y+Y13161 complex
To Be Published
|
|
2IM8
 
 | | X-Ray Crystal Structure of Protein yppE from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR213. | | Descriptor: | Hypothetical protein yppE, PHOSPHATE ION | | Authors: | Kuzin, A.P, Shastry, R, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Hang, D, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-03 | | Release date: | 2006-10-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray structure of hypothetical protein yPPE. Northeast Structural Genomics Consortium target SR213.
To be Published
|
|
4K6L
 
 | | Structure of Typhoid Toxin | | Descriptor: | Cytolethal distending toxin subunit B homolog, GLYCEROL, Putative pertussis-like toxin subunit | | Authors: | Gao, X, Song, J, Galan, J. | | Deposit date: | 2013-04-16 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin.
Nature, 499, 2013
|
|
4F9Z
 
 | | Crystal Structure of human ERp27 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Kober, F.X, Koelmel, W, Kuper, J, Schindelin, H. | | Deposit date: | 2012-05-21 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Protein-Disulfide Isomerase Family Member ERp27 Provides Insights into Its Substrate Binding Capabilities.
J.Biol.Chem., 288, 2013
|
|
5W7T
 
 | |
2UP1
 
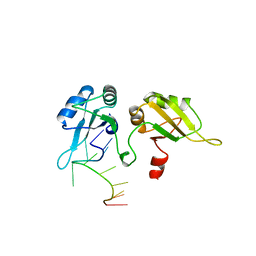 | | STRUCTURE OF UP1-TELOMERIC DNA COMPLEX | | Descriptor: | DNA (5'-D(P*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), PROTEIN (HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1) | | Authors: | Ding, J, Hayashi, M.K, Krainer, A.R, Xu, R.-M. | | Deposit date: | 1998-07-10 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA.
Genes Dev., 13, 1999
|
|
8TBY
 
 | | Apo Bcs1, unsymmetrized | | Descriptor: | Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-06-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8WPM
 
 | |
8TPL
 
 | | ATP-2 state of Bcs1 (unsymmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TP1
 
 | | ATP-2 state of Bcs1 (C7 symmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-08-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8TI0
 
 | | ATP-1 state of Bcs1 (unsymmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-07-18 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
2SPG
 
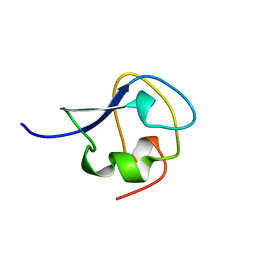 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15S | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-21 | | Release date: | 1999-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
