1EM8
 
 | | Crystal structure of chi and psi subunit heterodimer from DNA POL III | | Descriptor: | DNA POLYMERASE III CHI SUBUNIT, DNA POLYMERASE III PSI SUBUNIT | | Authors: | Gulbis, J.M, Finkelstein, J, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2000-03-16 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the chi:psi sub-assembly of the Escherichia coli DNA polymerase clamp-loader complex.
Eur.J.Biochem., 271, 2004
|
|
2DM6
 
 | | Crystal structure of anti-configuration of indomethacin and leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complex | | Descriptor: | INDOMETHACIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent leukotriene B4 12-hydroxydehydrogenase, ... | | Authors: | Hori, T, Ishijima, J, Yokomizo, T, Ago, H, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-20 | | Release date: | 2006-11-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of anti-configuration of indomethacin and leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complex reveals the structural basis of broad spectrum indomethacin efficacy
J.Biochem.(Tokyo), 140, 2006
|
|
2Y52
 
 | | Crystal structure of E496A mutant of the box pathway encoded ALDH from Burkholderia xenovorans LB400 | | Descriptor: | ALDEHYDE DEHYDROGENASE (BOX PATHWAY), GLYCEROL | | Authors: | Bains, J, Leon, R, Temke, K.G, Boulanger, M.J. | | Deposit date: | 2011-01-11 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Elucidating the Reaction Mechanism of the Benzoate Oxidation Pathway Encoded Aldehyde Dehydrogenase from Burkholderia Xenovorans Lb400.
Protein Sci., 20, 2011
|
|
2DP7
 
 | | Crystal Structure of D(CGCGAATXCGCG) Where X is 5-(N-aminohexyl)carbamoyl-2'-deoxyuridine | | Descriptor: | (6-AMINOHEXYL)CARBAMIC ACID, DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DAP*DTP*DUP*DCP*DGP*DCP*DG)-3'), MAGNESIUM ION, ... | | Authors: | Juan, E.C.M, Kondo, J, Kurihara, T, Ito, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2006-05-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of DNA:DNA and DNA:RNA duplexes containing 5-(N-aminohexyl)carbamoyl-modified uracils reveal the basis for properties as antigene and antisense molecules
Nucleic Acids Res., 35, 2007
|
|
2DPD
 
 | |
5U17
 
 | |
1N94
 
 | | Aryl Tetrahydropyridine Inhbitors of Farnesyltransferase: Glycine, Phenylalanine and Histidine Derivates | | Descriptor: | 2-{(5-{[BUTYL-(2-CYCLOHEXYL-ETHYL)-AMINO]-METHYL}-2'-METHYL-BIPHENYL-2-CARBONYL)-AMINO]-4-METHYLSULFANYL-BUTYRIC ACID, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Connor, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: glycine, phenylalanine and histidine derivatives.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
2DQP
 
 | | Structural analyses of DNA:DNA and RNA:DNA duplexes containing 5-(N-aminohexyl)carbamoyl modified uridines | | Descriptor: | (6-AMINOHEXYL)CARBAMIC ACID, DNA (5'-D(*DCP*(OMU)P*DCP*DTP*(OMU)P*DCP*DTP*DTP*DC)-3'), RNA (5'-R(*GP*AP*AP*GP*AP*AP*GP*AP*G)-3') | | Authors: | Juan, E.C.M, Kondo, J, Ito, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2006-05-29 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of DNA:DNA and DNA:RNA duplexes containing 5-(N-aminohexyl)carbamoyl-modified uracils reveal the basis for properties as antigene and antisense molecules
Nucleic Acids Res., 35, 2007
|
|
6MOM
 
 | | Crystal structure of human Interleukin-1 receptor associated Kinase 4 (IRAK 4, CID 100300) in complex with compound NCC00371481 (BSI 107591) | | Descriptor: | 1,2-ETHANEDIOL, 6-[7-methoxy-6-(1-methyl-1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-3-yl]-N-[(3R)-pyrrolidin-3-yl]pyridin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Abendroth, J, Mayclin, S.J, Lorimer, D.D, Starczynowski, D, Hoyt, S, Tawa, G, Thomas, C. | | Deposit date: | 2018-10-04 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Overcoming adaptive therapy resistance in AML by targeting immune response pathways.
Sci Transl Med, 11, 2019
|
|
2XF1
 
 | | Crystal structure of Plasmodium falciparum actin depolymerization factor 1 | | Descriptor: | COFILIN ACTIN-DEPOLYMERIZING FACTOR HOMOLOG 1, SULFATE ION | | Authors: | Singh, B.K, Sattler, J.M, Huttu, J, Chatterjee, M, Schueler, H, Kursula, I. | | Deposit date: | 2010-05-20 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures Explain Functional Differences in the Two Actin Depolymerization Factors of the Malaria Parasite.
J.Biol.Chem., 286, 2011
|
|
5U2V
 
 | |
2DSZ
 
 | | Three dimensional structure of a goat signalling protein secreted during involution | | Descriptor: | Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, J, Ethayathulla, A.S, Singh, N, Ujwal, R, Srivastava, D.B, Sharma, S, Singh, T.P. | | Deposit date: | 2006-07-09 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Carbohydrate-binding properties of goat secretory glycoprotein (SPG-40) and its functional implications: structures of the native glycoprotein and its four complexes with chitin-like oligosaccharides
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
1FFK
 
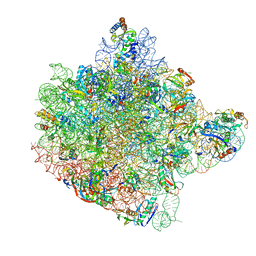 | | CRYSTAL STRUCTURE OF THE LARGE RIBOSOMAL SUBUNIT FROM HALOARCULA MARISMORTUI AT 2.4 ANGSTROM RESOLUTION | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Ban, N, Nissen, P, Hansen, J, Moore, P.B, Steitz, T.A. | | Deposit date: | 2000-07-25 | | Release date: | 2000-08-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The complete atomic structure of the large ribosomal subunit at 2.4 A resolution.
Science, 289, 2000
|
|
2DT0
 
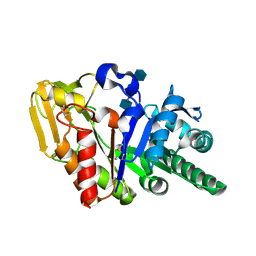 | | Crystal structure of the complex of goat signalling protein with the trimer of N-acetylglucosamine at 2.45A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, J, Ethayathulla, A.S, Srivastava, D.B, Singh, N, Sharma, S, Bhushan, A, Srinivasan, A, Singh, T.P. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Carbohydrate-binding properties of goat secretory glycoprotein (SPG-40) and its functional implications: structures of the native glycoprotein and its four complexes with chitin-like oligosaccharides
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
6MQL
 
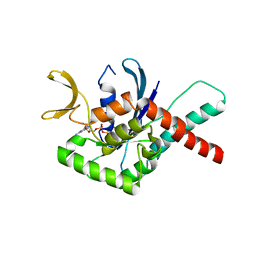 | | Crystal Structure of GTPase Domain of Human Septin 12 Mutant T89M | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Septin-12 | | Authors: | Castro, D.K.S.V, Pereira, H.M, Brandao-Neto, J, Ulian, A.P.U, Garratt, R.C. | | Deposit date: | 2018-10-10 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structure of GTPase Domain of Human Septin 12
To Be Published
|
|
6MJW
 
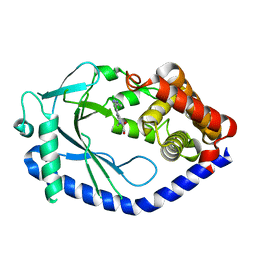 | | human cGAS catalytic domain bound with the inhibitor G150 | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Lama, L, Adura, C, Xie, W, Tomita, D, Kamei, T, Kuryavyi, V, Gogakos, T, Steinberg, J.I, Miller, M, Ramos-Espiritu, L, Asano, Y, Hashizume, S, Aida, J, Imaeda, T, Okamoto, R, Jennings, A.J, Michinom, M, Kuroita, T, Stamford, A, Gao, P, Meinke, P, Glickman, J.F, Patel, D.J, Tuschl, T. | | Deposit date: | 2018-09-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression.
Nat Commun, 10, 2019
|
|
1I5U
 
 | | SOLUTION STRUCTURE OF CYTOCHROME B5 TRIPLE MUTANT (E48A/E56A/D60A) | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qian, C, Yao, Y, Tang, W, Wang, J, Zhongxian, H. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of charged amino-acid mutation on the solution structure of cytochrome b(5) and binding between cytochrome b(5) and cytochrome c.
Protein Sci., 10, 2001
|
|
2XDP
 
 | | Crystal structure of the tudor domain of human JMJD2C | | Descriptor: | LYSINE-SPECIFIC DEMETHYLASE 4C, SULFATE ION | | Authors: | Yue, W.W, Gileadi, C, Krojer, T, Weisbach, H, Ugochukwu, E, Daniel, M, Phillips, C, Chaikuad, A, von Delft, F, Allerston, C, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of the Tudor Domain of Human Jmjd2C
To be Published
|
|
2DEM
 
 | | Crystal structure of Uracil-DNA glycosylase in complex with AP:A containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*AP*GP*CP*AP*AP*CP*A)-3', DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Hoseki, J, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
1N77
 
 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with tRNA(Glu) and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glutamyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-13 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
4IIF
 
 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with castanospermine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
2J67
 
 | | The TIR domain of human Toll-Like Receptor 10 (TLR10) | | Descriptor: | TOLL LIKE RECEPTOR 10 | | Authors: | Stenmark, P, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kotenyova, T, Magnusdottir, A, Nilsson, M.E, Nilsson-Ehle, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2006-09-26 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Human Toll-Like Receptor 10 Cytoplasmic Domain Reveals a Putative Signaling Dimer.
J.Biol.Chem., 283, 2008
|
|
2XOF
 
 | | Ribonucleotide reductase Y122NO2Y modified R2 subunit of E. coli | | Descriptor: | MU-OXO-DIIRON, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Yokoyama, K, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-08-15 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Hot Oxidant, 3-No(2)Y(122) Radical, Unmasks Conformational Gating in Ribonucleotide Reductase.
J.Am.Chem.Soc., 132, 2010
|
|
1N9A
 
 | | Farnesyltransferase complex with tetrahydropyridine inhibitors | | Descriptor: | 1-{2-[3-(4-CYANO-BENZYL)-3H-IMIDAZOL-4-YL]-ACETYL}-5-NAPHTHALEN-1-YL-1,2,3,6-TETRAHYDRO-PYRIDINE-4-CARBONITRILE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Conner, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: bioavailable analogues with improved cellular potency.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
6MOW
 
 | |
