4FLV
 
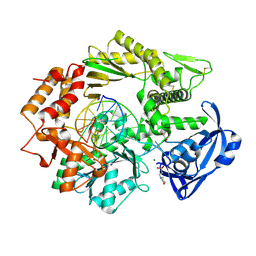 | |
4FMU
 
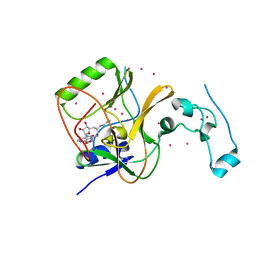 | | Crystal structure of Methyltransferase domain of human SET domain-containing protein 2 Compound: Pr-SNF | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-(propylamino)hexanoic acid, Histone-lysine N-methyltransferase SETD2, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Zeng, H, Ibanez, G, Zheng, W, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-18 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sinefungin Derivatives as Inhibitors and Structure Probes of Protein Lysine Methyltransferase SETD2.
J.Am.Chem.Soc., 134, 2012
|
|
4OBS
 
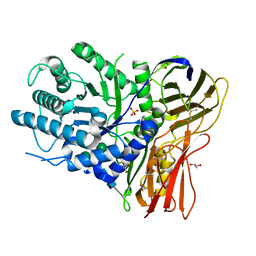 | | Crystal structure of human alpha-L-iduronidase in the P212121 form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, CHLORIDE ION, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, James, M.N.G. | | Deposit date: | 2014-01-07 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of human alpha-L-iduronidase in the P212121 form
To be Published
|
|
4OCW
 
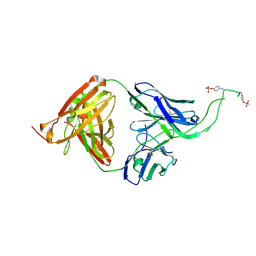 | | Crystal structure of human Fab CAP256-VRC26.06, a potent V1V2-directed HIV-1 neutralizing antibody | | Descriptor: | CAP256-VRC26.06 heavy chain, CAP256-VRC26.06 light chain | | Authors: | Gorman, J, Doria-Rose, N.A, Schramm, C.A, Moore, P.L, Mascola, J.R, Shapiro, L, Morris, L, Kwong, P.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies.
Nature, 509, 2014
|
|
4FC6
 
 | | Studies on DCR shed new light on peroxisomal beta-oxidation: Crystal structure of the ternary complex of pDCR | | Descriptor: | HEXANOYL-COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Peroxisomal 2,4-dienoyl-CoA reductase | | Authors: | Hua, T, Wu, D, Wang, J, Shaw, N, Liu, Z.-J. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal beta-oxidation of unsaturated fatty acids
J.Biol.Chem., 287, 2012
|
|
4FD5
 
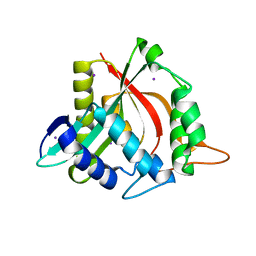 | |
4OM8
 
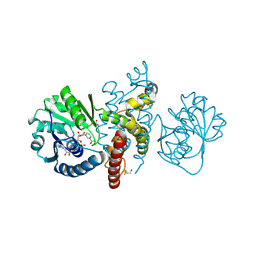 | | Crystal structure of 5-formly-3-hydroxy-2-methylpyridine 4-carboxylic acid (FHMPC) 5-dehydrogenase, an NAD+ dependent dismutase. | | Descriptor: | 3-hydroxybutyryl-coA dehydrogenase, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Mugo, A.N, Kobayashi, J, Mikami, B, Yagi, T, Ohnishi, K. | | Deposit date: | 2014-01-27 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylic acid 5-dehydrogenase, an NAD(+)-dependent dismutase from Mesorhizobium loti
Biochem.Biophys.Res.Commun., 456, 2015
|
|
4OEP
 
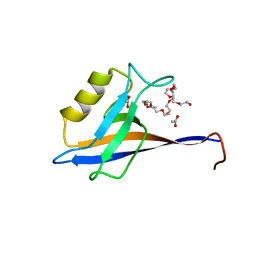 | |
4FD4
 
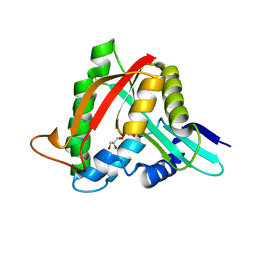 | |
4OO4
 
 | |
4OPN
 
 | |
8GOU
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH003 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH003 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
6Q8W
 
 | | Respiratory complex I from Thermus thermophilus with bound Aureothin. | | Descriptor: | Aureothin, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-12-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
1MD7
 
 | | Monomeric structure of the zymogen of complement protease C1r | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1R COMPLEMENT SERINE PROTEASE | | Authors: | Budayova-Spano, M, Grabarse, W, Thielens, N.M, Hillen, H, Lacroix, M, Schmidt, M, Fontecilla-Camps, J, Arlaud, G.J, Gaboriaud, C. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-07 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Monomeric structures of the zymogen and active catalytic domain of complement protease c1r: further insights into the c1 activation mechanism
Structure, 10, 2002
|
|
6QF9
 
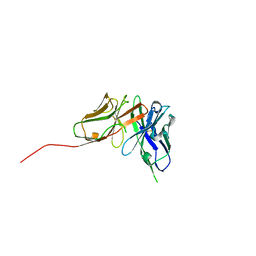 | | Structure of an anti-Mcl1 scFv | | Descriptor: | scFv | | Authors: | Luptak, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-11-06 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1MLY
 
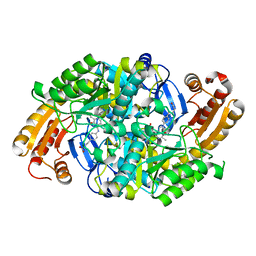 | | Crystal Structure of 7,8-Diaminopelargonic Acid Synthase in complex with the cis isomer of amiclenomycin | | Descriptor: | 7,8-diamino-pelargonic acid aminotransferase, CIS-AMICLENOMYCIN, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Sandmark, J, Mann, S, Marquet, A, Schneider, G. | | Deposit date: | 2002-09-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for the inhibition of the biosynthesis of biotin by the antibiotic amiclenomycin
J.Biol.Chem., 277, 2002
|
|
6QCZ
 
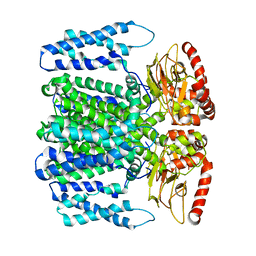 | | MloK1 model from single particle analysis of 2D crystals, class 3 (intermediate extended conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
6QD3
 
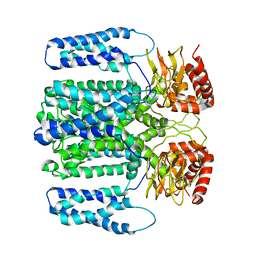 | | MloK1 model from single particle analysis of 2D crystals, class 7 (intermediate conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
1MP5
 
 | | Y177F VARIANT OF S. ENTERICA RmlA | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, Y177F VARIANT OF S. ENTERICA RmlA BOUND TO UDP-GLUCOSE | | Authors: | Barton, W.A, Biggins, J.B, Jiang, J, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2002-09-11 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6QF6
 
 | |
6QJP
 
 | | Cryo-EM structure of heparin-induced 2N4R tau jagged filaments | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Falcon, B, Murzin, A.G, Fan, J, Crowther, R.A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases.
Elife, 8, 2019
|
|
1MMU
 
 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-glucose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, beta-D-glucopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
1M1L
 
 | | Human Suppressor of Fused (N-terminal domain) | | Descriptor: | Suppressor of Fused | | Authors: | Merchant, M, Vajdos, F.F, Ultsch, M, Maun, H.R, Wendt, U, Cannon, J, Lazarus, R.A, de Vos, A.M, de Sauvage, F.J. | | Deposit date: | 2002-06-19 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Suppressor of fused regulates Gli activity through a dual binding mechanism
Mol.Cell.Biol., 24, 2004
|
|
1M20
 
 | | Crystal Structure of F35Y Mutant of Trypsin-solubilized Fragment of Cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yao, P, Wu, J, Wang, Y.-H, Sun, B.-Y, Xia, Z.-X, Huang, Z.-X. | | Deposit date: | 2002-06-20 | | Release date: | 2002-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallography, CD and kinetic studies revealed the essence of the abnormal behaviors of the cytochrome b5 Phe35-->Tyr mutant.
Eur.J.Biochem., 269, 2002
|
|
1M3K
 
 | | biosynthetic thiolase, inactive C89A mutant | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-28 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
