6KJ9
 
 | | E. coli ATCase catalytic subunit mutant - G128/130A | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
3X12
 
 | | Crystal structure of HLA-B*57:01.I80N | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-57 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2014-10-24 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The interaction of KIR3DL1*001 with HLA class I molecules is dependent upon molecular microarchitecture within the Bw4 epitope
J.Immunol., 194, 2015
|
|
6KKU
 
 | | human KCC1 structure determined in NaCl and GDN | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
5Z1V
 
 | | Crystal structure of AvrPib | | Descriptor: | AvrPib protein | | Authors: | Zhang, X, He, D, Zhao, Y.X, Taylor, I.A, Peng, Y.L, Yang, J, Liu, J.F. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | A positive-charged patch and stabilized hydrophobic core are essential for avirulence function of AvrPib in the rice blast fungus.
Plant J., 96, 2018
|
|
6KJ8
 
 | | E. coli ATCase holoenzyme mutant - G166P (catalytic chain) | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
4PJ7
 
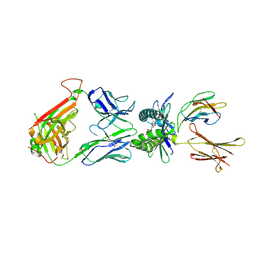 | | Structure of human MR1-5-OP-RU in complex with human MAIT TRBV6-4 TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
7XB3
 
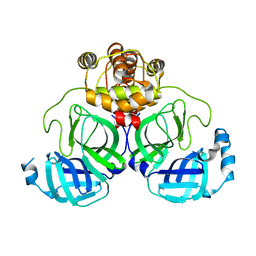 | |
3X0Y
 
 | | Crystal structure of FMN-bound DszC from Rhodococcus erythropolis D-1 | | Descriptor: | DszC, FLAVIN MONONUCLEOTIDE | | Authors: | Guan, L.J, Lee, W.C, Wang, S.P, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of apo-DszC and FMN-bound DszC from Rhodococcus erythropolis D-1.
Febs J., 282, 2015
|
|
3ZBI
 
 | | Fitting result in the O-layer of the subnanometer structure of the bacterial pKM101 type IV secretion system core complex digested with elastase | | Descriptor: | TRAF PROTEIN, TRAN PROTEIN, TRAO PROTEIN | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-10 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
7XAX
 
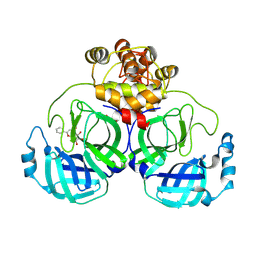 | |
7XB4
 
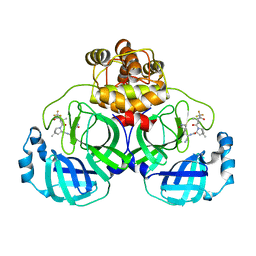 | | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Hu, X.H, Li, J, Zhang, J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with PF07321332
To Be Published
|
|
7X8U
 
 | |
1GHV
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-OXO-1,2-DIHYDRO-PYRIDIN-3-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, SODIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI9
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-5-METHOXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
4NNG
 
 | |
3X14
 
 | | Crystal structure of HLA-B*0801.N80I.R82L.G83R | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2014-10-25 | | Release date: | 2014-12-24 | | Last modified: | 2015-04-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The interaction of KIR3DL1*001 with HLA class I molecules is dependent upon molecular microarchitecture within the Bw4 epitope
J.Immunol., 194, 2015
|
|
4AO5
 
 | | B. subtilis prophage dUTPase YosS in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, SODIUM ION, SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEO TIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harrison, C, Turkenburg, J.P, Wilson, K.S. | | Deposit date: | 2012-03-23 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tying Down the Arm in Bacillus Dutpase: Structure and Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7X2N
 
 | | Crystal structure of Diels-Alderase PycR1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-25 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of hetero-Diels-Alderase PycR1
To Be Published
|
|
1GI2
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-3H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1NSG
 
 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-RAPAMYCIN COMPLEX INTERACTING WITH HUMAN FRAP | | Descriptor: | C49-METHYL RAPAMYCIN, FK506-BINDING PROTEIN, FKBP-RAPAMYCIN ASSOCIATED PROTEIN (FRAP) | | Authors: | Liang, J, Choi, J, Clardy, J. | | Deposit date: | 1997-07-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7X2S
 
 | | Crystal Structure of hetero-Diels-Alderase PycR1 in complex with Neosetophomone B and tropolone o-quinone methide | | Descriptor: | (1~{S},3~{R},11~{R},13~{S},14~{Z},18~{E})-1,8,14,17,17-pentamethyl-5,13-bis(oxidanyl)-2-oxatricyclo[9.9.0.0^{3,9}]icosa-4,7,9,14,18-pentaen-6-one, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Calcium-dependent glycosylated enzyme in the tandem hetero-Diels-Alder reaction
To Be Published
|
|
7X2X
 
 | | Crystal Structure of hetero-Diels-Alderase PycR1 in complex with 10-hydroxy-8E-humulene | | Descriptor: | (1R,2E,6E,9E)-2,5,5,9-tetramethylcycloundeca-2,6,9-trien-1-ol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Calcium-dependent glycosylated enzyme in the tandem hetero-Diels-Alder reaction
To Be Published
|
|
5Z2M
 
 | | Structure of Orp1L/Rab7 complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Oxysterol-binding protein-related protein 1, ... | | Authors: | Ma, X.L, Liu, K, Li, J, Li, H.H, Li, J, Yang, C.L, Liang, H.H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | A non-canonical GTPase interaction enables ORP1L-Rab7-RILP complex formation and late endosome positioning.
J. Biol. Chem., 293, 2018
|
|
7X36
 
 | | Crystal Structure of hetero-Diels-Alderase EupfF | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, J, Lu, J. | | Deposit date: | 2022-02-28 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Calcium-dependent glycosylated enzyme in the tandem hetero-Diels-Alder reaction
To Be Published
|
|
2PXJ
 
 | | The complex structure of JMJD2A and monomethylated H3K36 peptide | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, ... | | Authors: | Chen, Z, Zang, J, Kappler, J, Hong, X, Crawford, F, Zhang, G. | | Deposit date: | 2007-05-14 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the recognition of a methylated histone tail by JMJD2A
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
