6OGJ
 
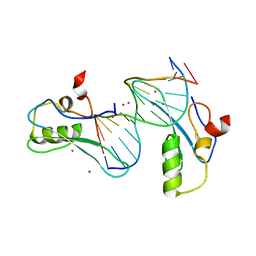 | | MeCP2 MBD in complex with DNA | | 分子名称: | DNA (5'-D(*CP*GP*GP*AP*GP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*CP*AP*CP*TP*CP*CP*G)-3'), Methyl-CpG-binding protein 2, ... | | 著者 | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2019-04-02 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
8A8W
 
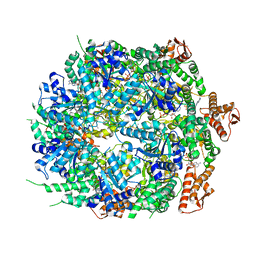 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Ecumycin (class 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.29 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
1KJM
 
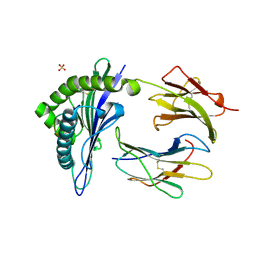 | | TAP-A-associated rat MHC class I molecule | | 分子名称: | B6 Peptide, RT1 class I histocompatibility antigen, AA alpha chain, ... | | 著者 | Rudolph, M.G, Stevens, J, Speir, J.A, Trowsdale, J, Butcher, G.W, Joly, E, Wilson, I.A. | | 登録日 | 2001-12-04 | | 公開日 | 2002-12-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structures of two rat MHC class Ia (RT1-A) molecules that are associated differentially
with peptide transporter alleles TAP-A and TAP-B.
J.Mol.Biol., 324, 2002
|
|
4MN0
 
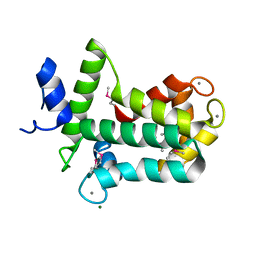 | | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca2+-loaded apoprotein conformation state | | 分子名称: | Berovin, CALCIUM ION, MAGNESIUM ION | | 著者 | Liu, Z.J, Stepanyuk, G.A, Vysotski, E.S, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2013-09-09 | | 公開日 | 2013-10-16 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca(2+)-loaded apoprotein conformation state.
Biochim.Biophys.Acta, 1834, 2013
|
|
7THH
 
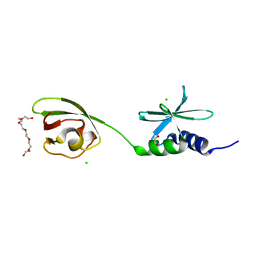 | | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | 著者 | Osipiuk, J, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-11 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein
to be published
|
|
8A8U
 
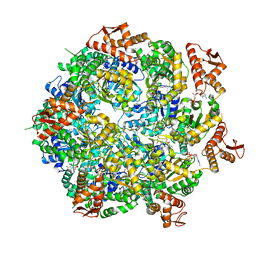 | | Mycobacterium tuberculosis ClpC1 hexamer structure | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
4HK5
 
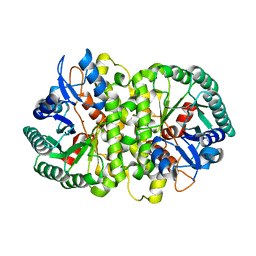 | | Crystal structure of Cordyceps militaris IDCase in apo form | | 分子名称: | Uracil-5-carboxylate decarboxylase, ZINC ION | | 著者 | Xu, S, Zhu, J, Ding, J. | | 登録日 | 2012-10-15 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
2X72
 
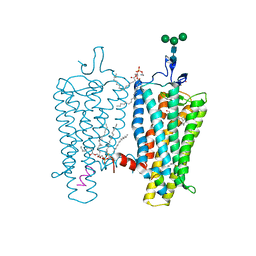 | | CRYSTAL STRUCTURE OF THE CONSTITUTIVELY ACTIVE E113Q,D2C,D282C RHODOPSIN MUTANT WITH BOUND GALPHACT PEPTIDE. | | 分子名称: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ACETATE ION, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | 著者 | Standfuss, J, Edwards, P.C, Dantona, A, Fransen, M, Xie, G, Oprian, D.D, Schertler, G.F.X. | | 登録日 | 2010-02-22 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Structural Basis of Agonist Induced Activation in Constitutively Active Rhodopsin
Nature, 471, 2011
|
|
4MJS
 
 | | crystal structure of a PB1 complex | | 分子名称: | 1,2-ETHANEDIOL, Protein kinase C zeta type, Sequestosome-1 | | 著者 | Ren, J, Wang, Z.X, Wu, J.W. | | 登録日 | 2013-09-04 | | 公開日 | 2014-08-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and biochemical insights into the homotypic PB1-PB1 complex between PKC zeta and p62
Sci China Life Sci, 57, 2014
|
|
8AIY
 
 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PAO1 IN COMPLEX WITH N-(beta-L-Fucopyranosyl)-biphenyl-3-carboxamide (4i) | | 分子名称: | CALCIUM ION, Fucose-binding lectin PA-IIL, N-(beta-L-Fucopyranosyl)-biphenyl-3-carboxamide, ... | | 著者 | Meiers, J, Mala, P, Varrot, A, Siebs, E, Imberty, A, Titz, A. | | 登録日 | 2022-07-27 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of N -beta-l-Fucosyl Amides as High-Affinity Ligands for the Pseudomonas aeruginosa Lectin LecB.
J.Med.Chem., 65, 2022
|
|
6OI9
 
 | | Crystal Structure of E. coli Biotin Carboxylase Complexed with 7-[3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine | | 分子名称: | 1,2-ETHANEDIOL, 7-[(3S)-3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | 著者 | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | 登録日 | 2019-04-09 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
4CNL
 
 | | Crystal structure of the Choline-binding domain of CbpL from Streptococcus pneumoniae | | 分子名称: | CHOLINE ION, GLYCEROL, PUTATIVE PNEUMOCOCCAL SURFACE PROTEIN, ... | | 著者 | Gutierrez-Fernandez, J, Bartual, S.G, Hermoso, J.A. | | 登録日 | 2014-01-23 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Modular Architecture and Unique Teichoic Acid Recognition Features of Choline-Binding Protein L (Cbpl) Contributing to Pneumococcal Pathogenesis.
Sci.Rep., 6, 2016
|
|
3FBW
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant C176Y | | 分子名称: | BENZOIC ACID, CHLORIDE ION, Haloalkane dehalogenase, ... | | 著者 | Dohnalek, J, Stsiapanava, A, Gavira, J.A, Kuta Smatanova, I, Kuty, M. | | 登録日 | 2008-11-20 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6OLU
 
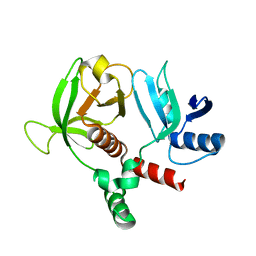 | | RIAM RA-PH core structure in the P212121 space group | | 分子名称: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | 著者 | Wu, J. | | 登録日 | 2019-04-17 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Phosphorylation of RIAM by src promotes integrin activation by unmasking the PH domain of RIAM.
Structure, 29, 2021
|
|
8B27
 
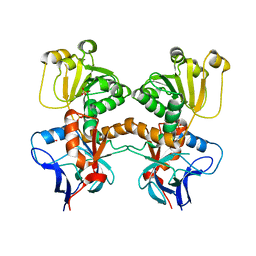 | | Dihydroprecondylocarpine acetate synthase from Catharanthus roseus | | 分子名称: | Dehydroprecondylocarpine acetate synthase, SULFATE ION | | 著者 | Langley, C, Basquin, J, Caputi, L, O'Connor, S.E. | | 登録日 | 2022-09-13 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
4HI7
 
 | | Crystal structure of glutathione transferase homolog from drosophilia mojavensis, TARGET EFI-501819, with bound glutathione | | 分子名称: | GI20122, GLUTATHIONE | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-10-11 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal structure of glutathione transferase homolog from drosophilia mojavensis, TARGET EFI-501819, with bound glutathione
To be Published
|
|
3FEA
 
 | |
4CYI
 
 | | Chaetomium thermophilum Pan3 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3-LIKE PROTEIN, ... | | 著者 | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | 登録日 | 2014-04-11 | | 公開日 | 2014-06-11 | | 最終更新日 | 2018-04-25 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation
Embo J., 33, 2014
|
|
7F43
 
 | |
3VQ7
 
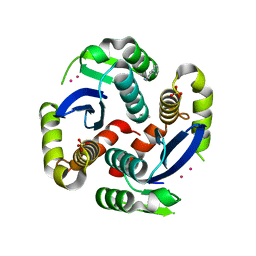 | | HIV-1 IN core domain in complex with 4-(1H-pyrrol-1-yl)aniline | | 分子名称: | 4-(1H-pyrrol-1-yl)aniline, CADMIUM ION, POL polyprotein, ... | | 著者 | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | 登録日 | 2012-03-20 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
4QF1
 
 | | Crystal structure of unliganded CH59UA, the inferred unmutated ancestor of the RV144 anti-HIV antibody lineage producing CH59 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CH59UA Fab fragment of heavy chain, CHLORIDE ION, ... | | 著者 | Wiehe, K, Easterhoff, D, Luo, K, Nicely, N.I, Bradley, T, Jaeger, F.H, Dennison, S.M, Zhang, R, Lloyd, K.E, Stolarchuk, C, Parks, R, Sutherland, L.L, Scearce, R.M, Morris, L, Kaewkungwal, J, Nitayaphan, S, Pitisuttithum, P, Rerks-Ngarm, S, Michael, N, Kim, J, Kelsoe, G, Montefiori, D.C, Tomaras, G, Bonsignori, M, Santra, S, Kepler, T.B, Alam, S.M, Moody, M.A, Liao, H.-X, Haynes, B.F. | | 登録日 | 2014-05-19 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Antibody Light-Chain-Restricted Recognition of the Site of Immune Pressure in the RV144 HIV-1 Vaccine Trial Is Phylogenetically Conserved.
Immunity, 41, 2014
|
|
3VSF
 
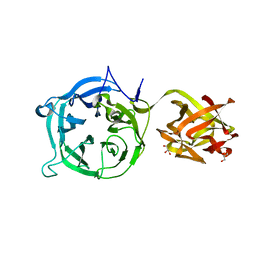 | | Crystal structure of 1,3Gal43A, an exo-beta-1,3-Galactanase from Clostridium thermocellum | | 分子名称: | GLYCEROL, Ricin B lectin | | 著者 | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | 登録日 | 2012-04-25 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.757 Å) | | 主引用文献 | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
5TKL
 
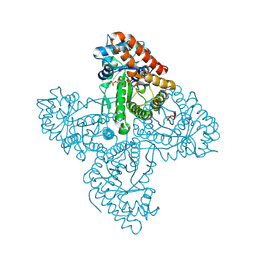 | | Crystal structure of FBP aldolase from Toxoplasma gondii, condensation intermediate | | 分子名称: | 1,3-DIHYDROXYACETONEPHOSPHATE, 1,6-di-O-phosphono-D-fructose, Fructose-bisphosphate aldolase, ... | | 著者 | Heron, P.W, Sygusch, J. | | 登録日 | 2016-10-06 | | 公開日 | 2017-10-04 | | 最終更新日 | 2020-01-08 | | 実験手法 | X-RAY DIFFRACTION (1.751 Å) | | 主引用文献 | Isomer activation controls stereospecificity of class I fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 292, 2017
|
|
7F42
 
 | |
7TOB
 
 | | Crystal structure of the SARS-CoV-2 Omicron main protease (Mpro) in complex with inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Sacco, M.D, Wang, J, Chen, Y. | | 登録日 | 2022-01-24 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition.
Cell Res., 32, 2022
|
|
