1V9Y
 
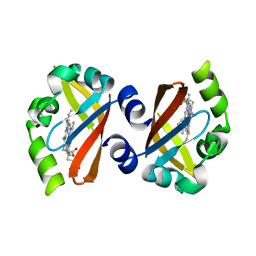 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (ferric form) | | Descriptor: | Heme pas sensor protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Lee, D.S, Watanabe, M, Sagami, I, Mikami, B, Raman, C.S, Shimizu, T. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor.
J.Biol.Chem., 279, 2004
|
|
5AWB
 
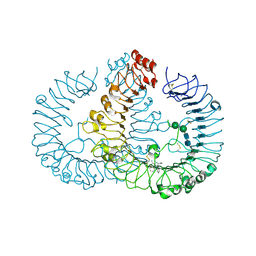 | | Crystal structure of human TLR8 in complex with N1-3-aminomethylbenzyl (meta-amine) | | Descriptor: | 1-[[3-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Human TLR8-Specific Agonists with Augmented Potency and Adjuvanticity.
J.Med.Chem., 58, 2015
|
|
5AWA
 
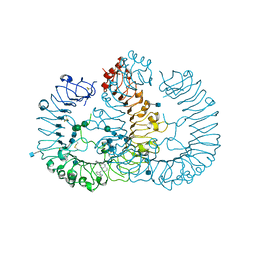 | | Crystal structure of human TLR8 in complex with MB-568 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[[3-(aminomethyl)phenyl]methyl]-3-pentyl-quinolin-2-amine, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2015-07-03 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based Design of Human TLR8-specific Agonists with Augmented Potency and Adjuvanticity
To Be Published
|
|
1V9Z
 
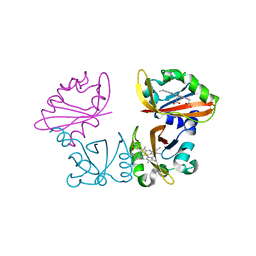 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (Ferrous Form) | | Descriptor: | Heme pas sensor protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Lee, D.S, Watanabe, M, Sagami, I, Mikami, B, Raman, C.S, Shimizu, T. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor.
J.Biol.Chem., 279, 2004
|
|
1VB6
 
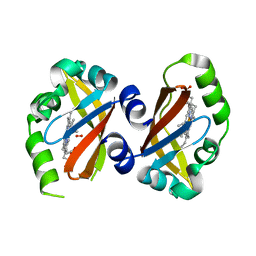 | | Crystal Structure of the heme PAS sensor domain of Ec DOS (oxygen-bound form) | | Descriptor: | Heme pas sensor protein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kurokawa, H, Watanabe, M, Sagami, I, Mikami, B, Shimizu, T. | | Deposit date: | 2004-02-24 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of oxygen-bound form of a Heme PAS domain of Ec DOS
To be Published
|
|
1J19
 
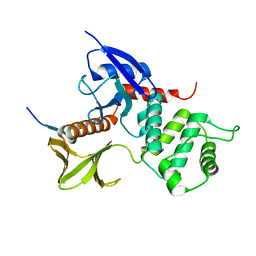 | | Crystal structure of the radxin FERM domain complexed with the ICAM-2 cytoplasmic peptide | | Descriptor: | 16-mer peptide from Intercellular adhesion molecule-2, radixin | | Authors: | Hamada, K, Shimizu, T, Yonemura, S, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex
EMBO J., 22, 2003
|
|
1NPM
 
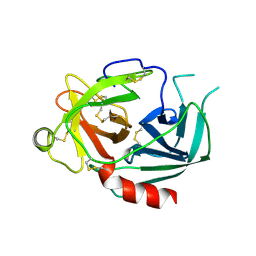 | | NEUROPSIN, A SERINE PROTEASE EXPRESSED IN THE LIMBIC SYSTEM OF MOUSE BRAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEUROPSIN | | Authors: | Kishi, T, Kato, M, Shimizu, T, Kato, K, Matsumoto, K, Yoshida, S, Shiosaka, S, Hakoshima, T. | | Deposit date: | 1998-01-07 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of neuropsin, a hippocampal protease involved in kindling epileptogenesis.
J.Biol.Chem., 274, 1999
|
|
1GLN
 
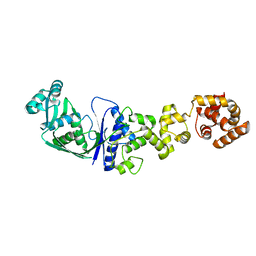 | | ARCHITECTURES OF CLASS-DEFINING AND SPECIFIC DOMAINS OF GLUTAMYL-TRNA SYNTHETASE | | Descriptor: | GLUTAMYL-TRNA SYNTHETASE | | Authors: | Nureki, O, Vassylyev, D.G, Katayanagi, K, Shimizu, T, Sekine, S, Kigawa, T, Miyazawa, T, Yokoyama, S, Morikawa, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1994-07-20 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architectures of class-defining and specific domains of glutamyl-tRNA synthetase.
Science, 267, 1995
|
|
1GC6
 
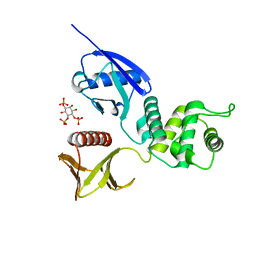 | | CRYSTAL STRUCTURE OF THE RADIXIN FERM DOMAIN COMPLEXED WITH INOSITOL-(1,4,5)-TRIPHOSPHATE | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, RADIXIN | | Authors: | Hamada, K, Shimizu, T, Matsui, T, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2000-07-21 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain.
EMBO J., 19, 2000
|
|
1GC7
 
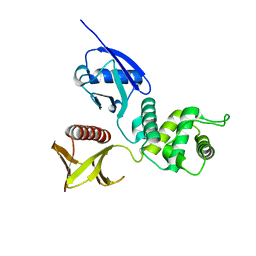 | | CRYSTAL STRUCTURE OF THE RADIXIN FERM DOMAIN | | Descriptor: | RADIXIN | | Authors: | Hamada, K, Shimizu, T, Matsui, T, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2000-07-21 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain.
EMBO J., 19, 2000
|
|
2IRF
 
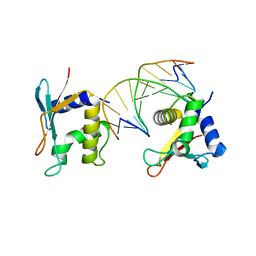 | | CRYSTAL STRUCTURE OF AN IRF-2/DNA COMPLEX. | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*TP*TP*TP*CP*AP*CP*(5IU)P*T)-3'), DNA (5'-D(P*AP*AP*GP*TP*GP*AP*AP*AP*GP*(5IU)P*GP*A)-3'), INTERFERON REGULATORY FACTOR 2, ... | | Authors: | Fujii, Y, Shimizu, T, Kusumoto, M, Kyogoku, Y, Taniguchi, T, Hakoshima, T. | | Deposit date: | 1999-05-30 | | Release date: | 1999-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences.
EMBO J., 18, 1999
|
|
3T6Q
 
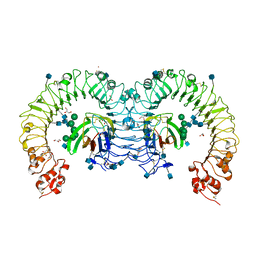 | | Crystal structure of mouse RP105/MD-1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD180 antigen, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Mouse and Human RP105/MD-1 Complexes Reveal Unique Dimer Organization of the Toll-Like Receptor Family.
J.Mol.Biol., 413, 2011
|
|
3THC
 
 | |
3THD
 
 | | Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2011-08-18 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of human beta-galactosidase: structural basis of Gm1 gangliosidosis and morquio B diseases
J.Biol.Chem., 287, 2012
|
|
2A0B
 
 | | HISTIDINE-CONTAINING PHOSPHOTRANSFER DOMAIN OF ARCB FROM ESCHERICHIA COLI | | Descriptor: | HPT DOMAIN, ZINC ION | | Authors: | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Refined structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor kinase ArcB from Escherichia coli at 1.57 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3VL8
 
 | | Crystal structure of XEG | | Descriptor: | SULFATE ION, Xyloglucan-specific endo-beta-1,4-glucanase A | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
3VL9
 
 | | Crystal structure of xeg-xyloglucan | | Descriptor: | Xyloglucan-specific endo-beta-1,4-glucanase A, beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
5DST
 
 | |
3VJI
 
 | | Human PPAR gamma ligand binding domain in complex with JKPL53 | | Descriptor: | (2S)-2-{4-butoxy-3-[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]benzoyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-10-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
3W3G
 
 | | Crystal structure of human TLR8 (unliganded form) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-21 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
3VJH
 
 | | Human PPAR GAMMA ligand binding domain in complex with JKPL35 | | Descriptor: | (2S)-2-[4-methoxy-3-({[4-(trifluoromethyl)benzoyl]amino}methyl)benzyl]pentanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-10-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
3VLA
 
 | | Crystal structure of edgp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EDGP | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
3VU7
 
 | | Crystal structure of REV1-REV7-REV3 ternary complex | | Descriptor: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Kikuchi, S, Hara, K, Shimizu, T, Sato, M, Hashimoto, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of recruitment of DNA polymerase [zeta] by interaction between REV1 and REV7 proteins
J.Biol.Chem., 287, 2012
|
|
3VLB
 
 | | Crystal structure of xeg-edgp | | Descriptor: | EDGP, Xyloglucan-specific endo-beta-1,4-glucanase A | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
3WPB
 
 | | Crystal structure of horse TLR9 (unliganded form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
