8HIN
 
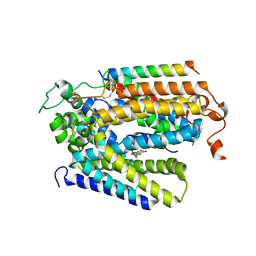 | | Structure of human SGLT2-MAP17 complex with Phlorizin | | Descriptor: | 1-[2-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,6-bis(oxidanyl)phenyl]-3-(4-hydroxyphenyl)propan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, PDZK1-interacting protein 1, ... | | Authors: | Hiraizumi, M, Kishida, H, Miyaguchi, I, Nureki, O. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8VC8
 
 | |
7ROX
 
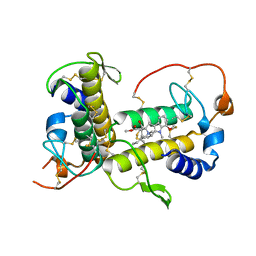 | | BthTX-I complexed with inhibitor MMV | | Descriptor: | 12-methoxy-Nb-methylvoachalotine, Basic phospholipase A2 homolog bothropstoxin-I | | Authors: | Borges, R.J, De Marino, I, Uson, I, Fontes, M.R.M. | | Deposit date: | 2021-08-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of a snake venom phospholipase A 2 -like protein complexed to an inhibitor from Tabernaemontana catharinensis.
Biochimie, 206, 2023
|
|
8J7R
 
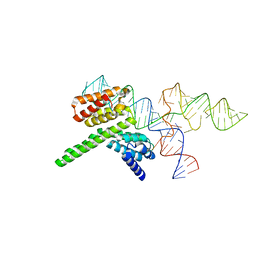 | | Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A (J-K-St/eIF4G focused) | | Descriptor: | Eukaryotic translation initiation factor 4 gamma 1, IRES RNA (J-K-St), MAGNESIUM ION | | Authors: | Suzuki, H, Fujiyoshi, Y, Imai, S, Shimada, I. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Dynamically regulated two-site interaction of viral RNA to capture host translation initiation factor.
Nat Commun, 14, 2023
|
|
8J3S
 
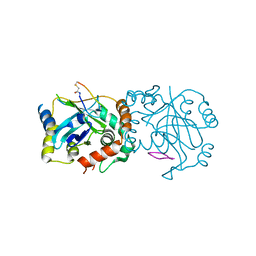 | | Complex structure of human cytomegalovirus protease and a macrocyclic peptide ligand | | Descriptor: | Assemblin, PHE-ILE-THR-GLY-HIS-TYR-TRP-VAL-ARG-PHE-LEU-PRO-CYS-GLY | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
8J3T
 
 | | Complex structure of human cytomegalovirus protease and a non-covalent small-molecule ligand | | Descriptor: | (4R)-1-[1-[(S)-[1-cyclopentyl-3-(2-methylphenyl)pyrazol-4-yl]-(4-methylphenyl)methyl]-2-oxidanylidene-pyridin-3-yl]-3-methyl-2-oxidanylidene-N-(3-oxidanylidene-2-azabicyclo[2.2.2]octan-4-yl)imidazolidine-4-carboxamide, Assemblin | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
8JMR
 
 | | Crystal structure of hinokiresinol synthase in complex with 1,7-bis(4-hydroxyphenyl)hepta-1,6-dien-3-one | | Descriptor: | 1,7-bis(4-hydroxyphenyl)hepta-1,6-dien-3-one, Hinokiresinol synthase alpha subunit, Hinokiresinol synthase beta subunit, ... | | Authors: | Ding, Y, Ushimaru, R, Mori, T, Abe, I. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insights into the C-C Bond-Forming Rearrangement Reaction Catalyzed by Heterodimeric Hinokiresinol Synthase.
J.Am.Chem.Soc., 145, 2023
|
|
8JMQ
 
 | | Crystal structure of hinokiresinol synthase | | Descriptor: | Hinokiresinol synthase alpha subunit, Hinokiresinol synthase beta subunit | | Authors: | Ding, Y, Ushimaru, R, Mori, T, Abe, I. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into the C-C Bond-Forming Rearrangement Reaction Catalyzed by Heterodimeric Hinokiresinol Synthase.
J.Am.Chem.Soc., 145, 2023
|
|
8BIZ
 
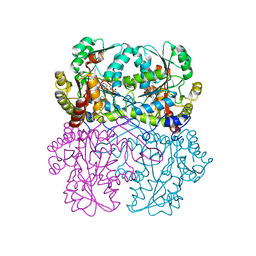 | | Cystathionine gamma-lyase from Toxoplasma gondii in complex with cysteine | | Descriptor: | CYSTEINE, Cystathionine beta-lyase, putative, ... | | Authors: | Fernandez-Rodriguez, C, Conter, C, Oyenarte, I, Favretto, F, Quintana, I, Martinez-Chantar, M.L, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2022-11-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Structural basis of the inhibition of cystathionine gamma-lyase from Toxoplasma gondii by propargylglycine and cysteine.
Protein Sci., 32, 2023
|
|
8BIW
 
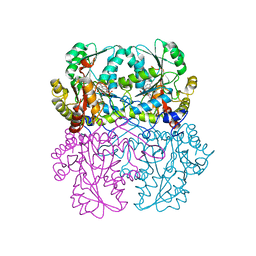 | | Cystathionine gamma-lyase N360S mutant in complex with DL-propargylglycine | | Descriptor: | (2S)-2-aminopent-4-enoic acid, Cystathionine beta-lyase, putative, ... | | Authors: | Fernandez-Rodriguez, C, Conter, C, Oyenarte, I, Favretto, F, Quintana, I, Martinez-Chantar, M.L, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2022-11-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Structural basis of the inhibition of cystathionine gamma-lyase from Toxoplasma gondii by propargylglycine and cysteine.
Protein Sci., 32, 2023
|
|
8BIU
 
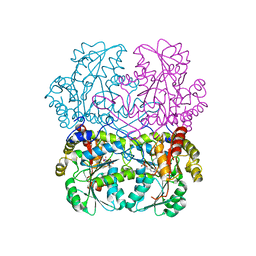 | | Cystathionine gamma-lyase in complex with cystathionine | | Descriptor: | 2-KETOBUTYRIC ACID, Cystathionine beta-lyase, putative, ... | | Authors: | Fernandez-Rodriguez, C, Conter, C, Oyenarte, I, Favretto, F, Quintana, I, Martinez-Chantar, M.L, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2022-11-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural basis of the inhibition of cystathionine gamma-lyase from Toxoplasma gondii by propargylglycine and cysteine.
Protein Sci., 32, 2023
|
|
8BIS
 
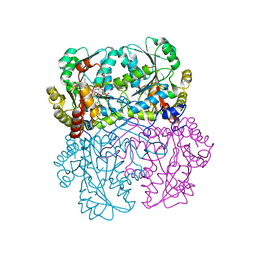 | | Crystal structure of cystathionine gamma-lyase from Toxoplasma gondii in complex with DL-propargylglycine | | Descriptor: | (2S)-2-aminopent-4-enoic acid, Cystathionine beta-lyase, putative, ... | | Authors: | Fernandez-Rodriguez, C, Conter, C, Oyenarte, I, Favretto, F, Quintana, I, Martinez-Chantar, M.L, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2022-11-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.266 Å) | | Cite: | Structural basis of the inhibition of cystathionine gamma-lyase from Toxoplasma gondii by propargylglycine and cysteine.
Protein Sci., 32, 2023
|
|
8BIV
 
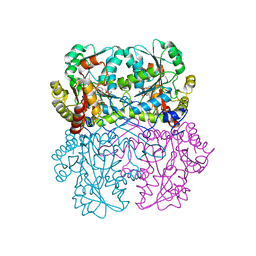 | | Cystathionine gamma-lyase N360S mutant from Toxoplasma gondii | | Descriptor: | Cystathionine beta-lyase, putative, GLYCEROL, ... | | Authors: | Fernandez-Rodriguez, C, Conter, C, Oyenarte, I, Favretto, F, Quintana, I, Martinez-Chantar, M.L, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2022-11-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis of the inhibition of cystathionine gamma-lyase from Toxoplasma gondii by propargylglycine and cysteine.
Protein Sci., 32, 2023
|
|
8BIX
 
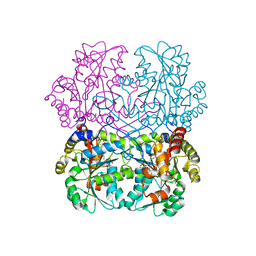 | | Cystathionine gamma-lyase N360S mutant from Toxoplasma gondii in complex with cystathionine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Cystathionine beta-lyase, putative, ... | | Authors: | Fernandez-Rodriguez, C, Conter, C, Oyenarte, I, Favretto, F, Quintana, I, Martinez-Chantar, M.L, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2022-11-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis of the inhibition of cystathionine gamma-lyase from Toxoplasma gondii by propargylglycine and cysteine.
Protein Sci., 32, 2023
|
|
6M48
 
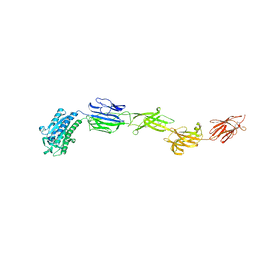 | | Crystal structure of pilus adhesin, SpaC from Lactobacillus rhamnosus GG - P21212 form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SpaC | | Authors: | Kant, A, Palva, A, Von Ossowaski, I, Krishnan, V. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
5E3B
 
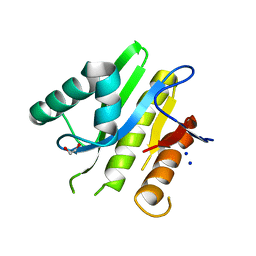 | | Structure of macrodomain protein from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, Macrodomain protein, SODIUM ION | | Authors: | Lalic, J, Posavec Marjanovic, M, Perina, D, Sabljic, I, Zaja, R, Plese, B, Imesek, M, Bucca, G, Ahel, M, Cetkovic, H, Luic, M, Mikoc, A, Ahel, I. | | Deposit date: | 2015-10-02 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Disruption of Macrodomain Protein SCO6735 Increases Antibiotic Production in Streptomyces coelicolor.
J.Biol.Chem., 291, 2016
|
|
8CP6
 
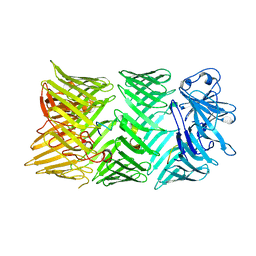 | |
8CR1
 
 | | Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1,GET2-GET1, ZINC ION | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
4Y6M
 
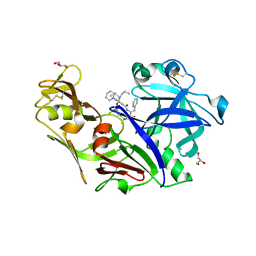 | | Structure of plasmepsin II from Plasmodium falciparum complexed with inhibitor PG418 | | Descriptor: | GLYCEROL, Plasmepsin-2, ~{N}1-[(~{Z},3~{R})-4-[2-(3-methoxyphenyl)propan-2-ylamino]-3-oxidanyl-1-phenyl-but-1-en-2-yl]-5-piperidin-1-yl-~{N}3,~{N}3-dipropyl-benzene-1,3-dicarboxamide | | Authors: | Recacha, R, Akopjana, I, Tars, K, Jaudzems, K. | | Deposit date: | 2015-02-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of plasmepsin II from Plasmodium falciparum in complex with two hydroxyethylamine-based inhibitors.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4ZDV
 
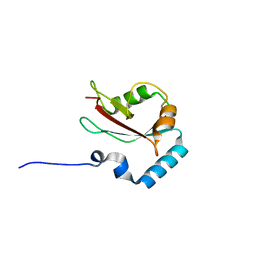 | |
4W5W
 
 | | Rubisco activase from Arabidopsis thaliana | | Descriptor: | Ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic, SULFATE ION | | Authors: | Hasse, D, Larsson, A.M, Andersson, I. | | Deposit date: | 2014-08-19 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Arabidopsis thaliana Rubisco activase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8R8D
 
 | |
8R05
 
 | | Photorhabdus lamondii ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.5 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R03
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R04
 
 | | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active Site Inhibitor Cystargolide A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
