4URF
 
 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, BICARBONATE ION, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
6D8X
 
 | | PPAR gamma LBD complexed with the agonist GW1929 | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, CITRATE ANION, GLYCEROL, ... | | Authors: | Mou, T.C, Chrisman, I.M, Hughes, T.S, Sprang, S.R. | | Deposit date: | 2018-04-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PPAR gamma LBD complexed with the agonist GW1929
To Be Published
|
|
6DB1
 
 | | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica | | Descriptor: | CHLORIDE ION, Putative methyl-accepting chemotaxis protein | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica.
To Be Published
|
|
6D94
 
 | | Crystal structure of PPAR gamma in complex with Mediator of RNA polymerase II transcription subunit 1 | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Mediator of RNA polymerase II transcription subunit 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Mou, T.C, Chrisman, I.M, Hughes, T.S, Sprang, S.R. | | Deposit date: | 2018-04-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural mechanism of nuclear receptor biased agonism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6DBA
 
 | | Crystal Structure of VHH R303 | | Descriptor: | nanobody VHH R303 | | Authors: | Brooks, C.L, Toride King, M, Huh, I. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of VHH-mediated neutralization of the food-borne pathogenListeria monocytogenes.
J. Biol. Chem., 293, 2018
|
|
6DLL
 
 | | 2.2 Angstrom Resolution Crystal Structure of P-Hydroxybenzoate Hydroxylase from Pseudomonas putida in Complex with FAD. | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-01 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural comparison of p-hydroxybenzoate hydroxylase (PobA) from Pseudomonas putida with PobA from other Pseudomonas spp. and other monooxygenases.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6D7P
 
 | | Crystal Structure of Rat TRPV6*-Y466A | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Singh, A.K, Saotome, K, McGoldrick, L.L, Sobolevsky, A.I. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB.
Nat Commun, 9, 2018
|
|
4WQ7
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - "as isolated" form | | Descriptor: | HEME C, IODIDE ION, SULFATE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
4WQE
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - K208G mutant | | Descriptor: | HEME C, IODIDE ION, SULFATE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
4WFN
 
 | | Crystal structure of the large ribosomal subunit (50S) of Deinococcus radiodurans containing a three residue insertion in L22 in complex with erythromycin | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-09-16 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel.
Structure, 25, 2017
|
|
4WEQ
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in complex with NADP and sulfate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sroka, P, Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Osinski, T, Hillerich, B.S, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-10 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
6D7Q
 
 | | Crystal Structure of Rat TRPV6*-Y466A in complex with 2-Aminoethoxydiphenyl borate (2-APB) | | Descriptor: | 2-aminoethyl diphenylborinate, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, CALCIUM ION, ... | | Authors: | Singh, A.K, Saotome, K, McGoldrick, L.L, Sobolevsky, A.I. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.497 Å) | | Cite: | Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB.
Nat Commun, 9, 2018
|
|
6DSP
 
 | | LsrB from Clostridium saccharobutylicum in complex with AI-2 | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, Autoinducer 2-binding protein LsrB | | Authors: | Torcato, I.M, Xavier, K.B, Miller, S.T. | | Deposit date: | 2018-06-14 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Identification of novel autoinducer-2 receptors inClostridiareveals plasticity in the binding site of the LsrB receptor family.
J.Biol.Chem., 294, 2019
|
|
6D7X
 
 | | Crystal Structure of Rat TRPV6*-Y466A- in complex with brominated 2-Aminoethoxydiphenyl borate (2-APB-Br) | | Descriptor: | 2-aminoethyl (4-bromophenyl)phenylborinate, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, CALCIUM ION, ... | | Authors: | Singh, A.K, Saotome, K, McGoldrick, L.L, Sobolevsky, A.I. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB.
Nat Commun, 9, 2018
|
|
4WQD
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - K208G mutant | | Descriptor: | 1,2-ETHANEDIOL, GUANIDINE, HEME C, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
7OPB
 
 | | IL7R in complex with an antagonist | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IL7R binder, ... | | Authors: | Markovic, I, Verschueren, K.H.G, Verstraete, K, Savvides, S.N. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
4WFB
 
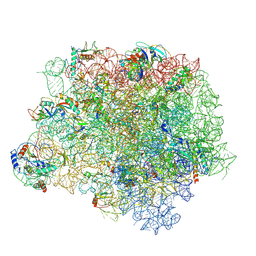 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with BC-3205 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S rRNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | Deposit date: | 2014-09-14 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WQA
 
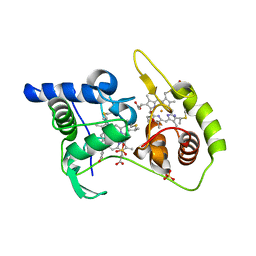 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - tetrathionate co-crystallization | | Descriptor: | 1,2-ETHANEDIOL, HEME C, IODIDE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
4WQ8
 
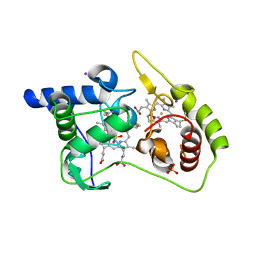 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - tetrathionate soak | | Descriptor: | HEME C, IODIDE ION, SULFATE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.3989 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
4WPH
 
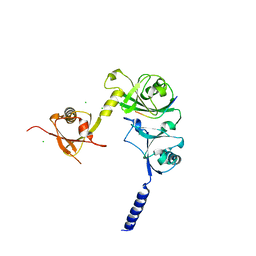 | |
7P7A
 
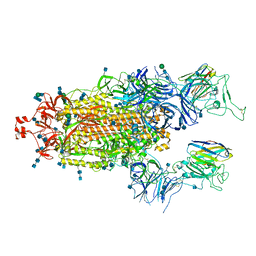 | | SARS-CoV-2 spike protein in complex with sybody#68 in a 2up/1flexible conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (4.76 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P78
 
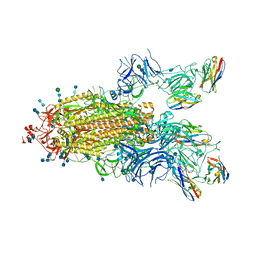 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 1up/1up-out/1down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P79
 
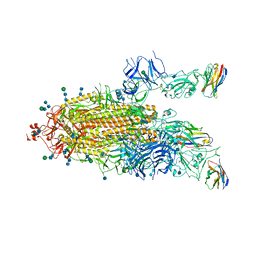 | | SARS-CoV-2 spike protein in complex with sybodyb#15 in a 1up/1up-out/1down conformation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
6O6C
 
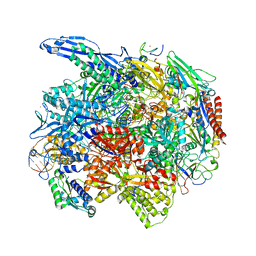 | | RNA polymerase II elongation complex arrested at a CPD lesion | | Descriptor: | DNA (27-MER), DNA (5'-D(P*GP*GP*AP*GP*AP*AP*GP*GP*AP*GP*CP*AP*GP*AP*GP*C)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Lahiri, I, Leshziner, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | 3.1 angstrom structure of yeast RNA polymerase II elongation complex stalled at a cyclobutane pyrimidine dimer lesion solved using streptavidin affinity grids.
J.Struct.Biol., 207, 2019
|
|
6O9L
 
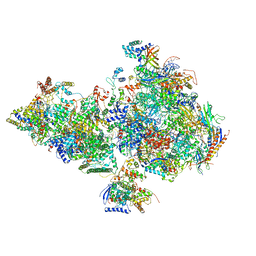 | | Human holo-PIC in the closed state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
