7AV4
 
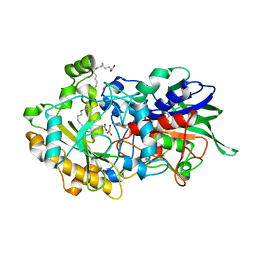 | | Dark state structure of the C432S mutant of Fatty Acid Photodecarboxylase (FAP) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Schlichting, I, Hartmann, E, Arnoux, P, Sorigue, D, Beisson, F. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
7AL0
 
 | | Crystal Structure of Heymonin, a Novel Frog-derived Peptide | | Descriptor: | CHLORIDE ION, Heymonin | | Authors: | Kascakova, B, Prudnikova, T, Kuta Smatanova, I, Xu, X. | | Deposit date: | 2020-10-03 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization and functional analysis of cathelicidin-MH, a novel frog-derived peptide with anti-septicemic properties.
Elife, 10, 2021
|
|
6F6I
 
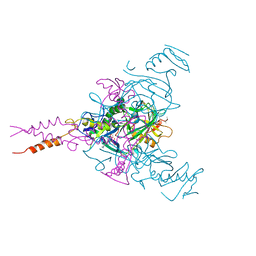 | | CRYSTAL STRUCTURE OF EBOLAVIRUS GLYCOPROTEIN IN COMPLEX WITH PAROXETINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Envelope glycoprotein, ... | | Authors: | Ren, J, Zhao, Y, Fry, E.E, Stuart, D.I. | | Deposit date: | 2017-12-05 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Target Identification and Mode of Action of Four Chemically Divergent Drugs against Ebolavirus Infection.
J. Med. Chem., 61, 2018
|
|
1J4Q
 
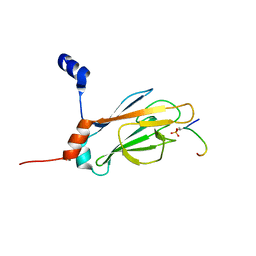 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T192) PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
6F8C
 
 | |
3LYR
 
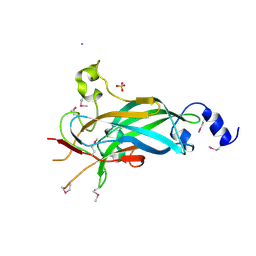 | | Human Early B-cell Factor 1 (EBF1) DNA-binding domain | | Descriptor: | AMMONIUM ION, SULFATE ION, Transcription factor COE1, ... | | Authors: | Siponen, M.I, Wisniewska, M, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kraulis, P, Kotenyova, T, Markova, N, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schuler, H, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-28 | | Release date: | 2010-03-16 | | Last modified: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Determination of Functional Domains in Early B-cell Factor (EBF) Family of Transcription Factors Reveals Similarities to Rel DNA-binding Proteins and a Novel Dimerization Motif.
J.Biol.Chem., 285, 2010
|
|
1W4Y
 
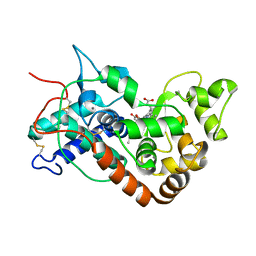 | | Ferrous horseradish peroxidase C1A in complex with carbon monoxide | | Descriptor: | CALCIUM ION, CARBON MONOXIDE, HORSERADISH PEROXIDASE C1A, ... | | Authors: | Carlsson, G.H, Nicholls, P, Svistunenko, D, Berglund, G.I, Hajdu, J. | | Deposit date: | 2004-08-03 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complexes of Horseradish Peroxidase with Formate, Acetate, and Carbon Monoxide
Biochemistry, 44, 2005
|
|
4S23
 
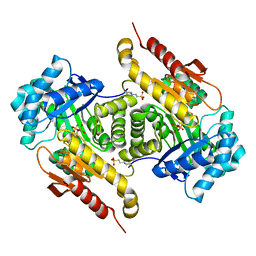 | | Structure of the GcpE-HMBPP complex from Thermus thermophilius | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Rekittke, I, Warkentin, E, Jomaa, H, Ermler, E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the GcpE-HMBPP complex from Thermus thermophilius.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
7AKD
 
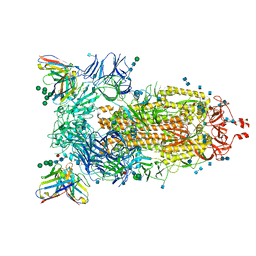 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the 47D11 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 47D11 neutralizing antibody heavy chain, ... | | Authors: | Fedry, J, Hurdiss, D.L, Wang, C, Li, W, Obal, G, Drulyte, I, Howes, S.C, van Kuppeveld, F.J.M, Foerster, F, Bosch, B.J. | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11.
Sci Adv, 7, 2021
|
|
4TJW
 
 | | Crystal Structure of human Tankyrase 2 in complex with PJ-34. | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TKG
 
 | | Crystal Structure of human Tankyrase 2 in complex with AZD2281. | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, GLYCEROL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TKS
 
 | |
6EUC
 
 | | Reactivating oxime bound to Tc AChE's catalytic gorge. | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-(5-morpholin-4-ylpentyl)pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | de la Mora, E, Weik, M, Braiki, A, Mougeot, R, Jean, L, Renard, P.I. | | Deposit date: | 2017-10-30 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.21998858 Å) | | Cite: | Potent 3-Hydroxy-2-Pyridine Aldoxime Reactivators of Organophosphate-Inhibited Cholinesterases with Predicted Blood-Brain Barrier Penetration.
Chemistry, 24, 2018
|
|
7MWW
 
 | | Structure of hepatitis C virus envelope full-length glycoprotein 2 (eE2) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, ... | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
4DM0
 
 | | TN5 transposase: 20MER OUTSIDE END 2 MN complex | | Descriptor: | 1,2-ETHANEDIOL, DNA NON-TRANSFERRED STRAND, DNA TRANSFERRED STRAND, ... | | Authors: | Klenchin, V.A, Lovell, S, Goryshin, I.Y, Reznikoff, W.R, Rayment, I. | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two-metal active site binding of a TN5 transposase synaptic complex
Nat.Struct.Biol., 9, 2002
|
|
2CJ9
 
 | | Crystal structure of Methanosarcina barkeri seryl-tRNA synthetase complexed with an analog of seryladenylate | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, CHLORIDE ION, SERYL-TRNA SYNTHETASE, ... | | Authors: | Bilokapic, S, Maier, T, Ahel, D, Gruic-Sovulj, I, Soll, D, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2006-03-29 | | Release date: | 2006-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Unusual Seryl-tRNA Synthetase Reveals a Distinct Zinc-Dependent Mode of Substrate Recognition
Embo J., 25, 2006
|
|
8DZZ
 
 | | Cryo-EM structure of chi dynein bound to Lis1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Reimer, J.M, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Lis1 relieves cytoplasmic dynein-1 autoinhibition by acting as a molecular wedge.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1D6J
 
 | |
6H0B
 
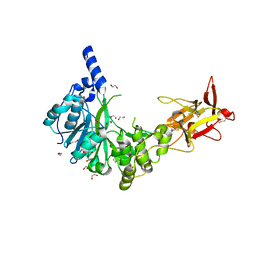 | | Crystal structure of the human GalNAc-T4 in complex with UDP, manganese and the diglycopeptide 6. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, ALA-THR-GLY-ALA-GLY-ALA-GLY-ALA-GLY-THR-THR-PRO-GLY-PRO-GLY, ... | | Authors: | de las Rivas, M, Daniel, E.J.P, Coelho, H, Lira-Navarrete, E, Raich, L, Companon, I, Diniz, A, Lagartera, L, Jimenez-Barbero, J, Clausen, H, Rovira, C, Marcelo, F, Corzana, F, Gerken, T.A, Hurtado-Guerrero, R. | | Deposit date: | 2018-07-08 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Insights into the Catalytic-Domain-Mediated Short-Range Glycosylation Preferences of GalNAc-T4.
ACS Cent Sci, 4, 2018
|
|
5CU5
 
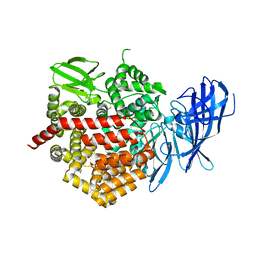 | | Crystal structure of ERAP2 without catalytic Zn(II) atom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Mathioudakis, N, Giastas, P, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural Basis for Antigenic Peptide Recognition and Processing by Endoplasmic Reticulum (ER) Aminopeptidase 2.
J.Biol.Chem., 290, 2015
|
|
1CK1
 
 | |
8ECZ
 
 | | Bovine Fab 4C1 | | Descriptor: | 4C1 Fab heavy chain, 4C1 Fab light chain, PHOSPHATE ION | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7NLI
 
 | |
2D24
 
 | | Crystal structure of ES complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
6H1C
 
 | | Structure of Ferric uptake regulator from Pseudomonas aeruginosa with manganese. | | Descriptor: | Ferric uptake regulation protein, MANGANESE (II) ION, ZINC ION | | Authors: | Nader, S, Perard, J, Carpentier, P, Michaud-Soret, I, Crouzy, S. | | Deposit date: | 2018-07-11 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ferric uptake regulator with Mn and Zn form Pseudomonas aeruginosa
To Be Published
|
|
