7AKJ
 
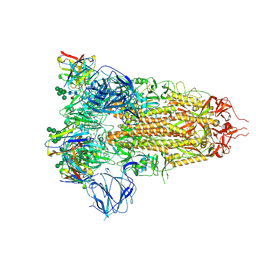 | | Structure of the SARS-CoV spike glycoprotein in complex with the 47D11 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fedry, J, Hurdiss, D.L, Wang, C, Li, W, Obal, G, Drulyte, I, Howes, S.C, van Kuppeveld, F.J.M, Foerster, F, Bosch, B.J. | | Deposit date: | 2020-10-01 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11.
Sci Adv, 7, 2021
|
|
7AKD
 
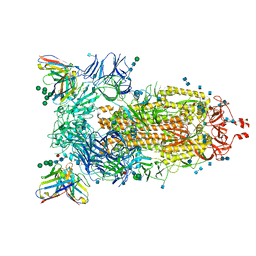 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the 47D11 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 47D11 neutralizing antibody heavy chain, ... | | Authors: | Fedry, J, Hurdiss, D.L, Wang, C, Li, W, Obal, G, Drulyte, I, Howes, S.C, van Kuppeveld, F.J.M, Foerster, F, Bosch, B.J. | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-19 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11.
Sci Adv, 7, 2021
|
|
4CJ1
 
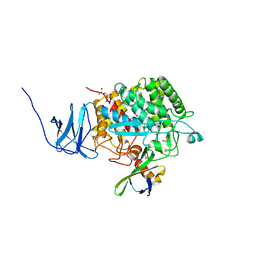 | | Crystal structure of CelD in complex with affitin H3 | | Descriptor: | CALCIUM ION, ENDOGLUCANASE D, GLYCEROL, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
4CJ0
 
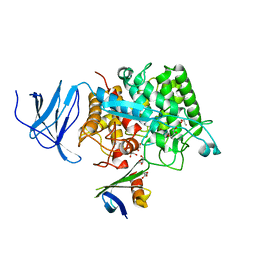 | | Crystal structure of CelD in complex with affitin E12 | | Descriptor: | CALCIUM ION, E12 AFFITIN, ENDOGLUCANASE D, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
4CJ2
 
 | | Crystal structure of HEWL in complex with affitin H4 | | Descriptor: | AFFITIN H4, GLYCEROL, LYSOZYME C | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
4PH2
 
 | | mature N-terminal domain of capsid protein from bovine leukemia virus | | Descriptor: | BLV capsid - N-terminal domain, GLYCEROL, SULFATE ION | | Authors: | Trajtenberg, F, Obal, G, Pritsch, O, Buschiazzo, A. | | Deposit date: | 2014-05-03 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | STRUCTURAL VIROLOGY. Conformational plasticity of a native retroviral capsid revealed by x-ray crystallography.
Science, 349, 2015
|
|
4PH3
 
 | | N-terminal domain of the capsid protein from bovine leukaemia virus (with no beta-hairpin) | | Descriptor: | BLV capsid, GLYCEROL, IODIDE ION | | Authors: | Trajtenberg, F, Obal, G, Pritsch, O, Buschiazzo, A. | | Deposit date: | 2014-05-03 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | STRUCTURAL VIROLOGY. Conformational plasticity of a native retroviral capsid revealed by x-ray crystallography.
Science, 349, 2015
|
|
4PH0
 
 | |
4PH1
 
 | |
3PJQ
 
 | | Trypanosoma cruzi trans-sialidase-like inactive isoform (including the natural mutation Tyr342His) in complex with lactose | | Descriptor: | Trans-sialidase, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Oppezzo, P, Baraibar, M, Obal, G, Pritsch, O, Alzari, P.M, Buschiazzo, A. | | Deposit date: | 2010-11-10 | | Release date: | 2011-06-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an enzymatically inactive trans-sialidase-like lectin from Trypanosoma cruzi: the carbohydrate binding mechanism involves residual sialidase activity.
Biochim.Biophys.Acta, 1814, 2011
|
|
