4UAC
 
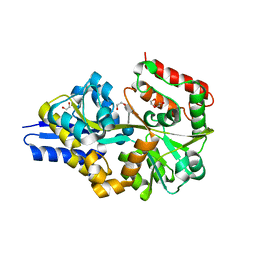 | | EUR_01830 with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Carbohydrate ABC transporter substrate-binding protein, ... | | Authors: | Koropatkin, N.M, Orlovsky, N.I. | | Deposit date: | 2014-08-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale.
Mol.Microbiol., 95, 2015
|
|
6TI1
 
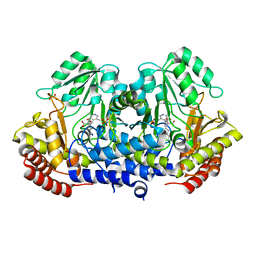 | | SHMT from Streptococcus thermophilus Tyr55Ser variant in complex with PLP/L-Threonine/Lys230 gem diamine complex | | Descriptor: | GLYCEROL, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-threonine, SODIUM ION, ... | | Authors: | Petrillo, G, Hernandez, K, Bujons, J, Clapes, P, Uson, I. | | Deposit date: | 2019-11-21 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into nucleophile substrate specificity in variants of N-Serine hydroxymethyltransferase from Streptococcus thermophilus
To Be Published
|
|
4UBF
 
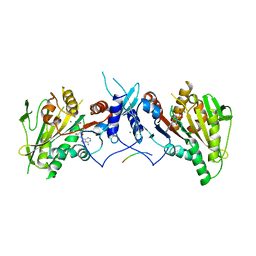 | | HsMCAK motor domain complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF2C, MAGNESIUM ION | | Authors: | Welburn, J.P.I, Talapatra, S.K. | | Deposit date: | 2014-08-12 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C-terminal region of the motor protein MCAK controls its structure and activity through a conformational switch.
Elife, 4, 2015
|
|
4UE5
 
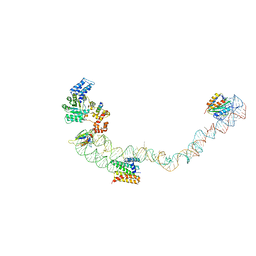 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | Descriptor: | 7S RNA, SIGNAL RECOGNITION PARTICLE 54 KDA PROTEIN, SIGNAL RECOGNITION PARTICLE 9 KDA PROTEIN, ... | | Authors: | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7O3B
 
 | |
6TG8
 
 | | Crystal structure of the Kelch domain in complex with 11 amino acid peptide (model of the ETGE loop) | | Descriptor: | Kelch-like ECH-associated protein 1, SODIUM ION, VAL-ILE-ASN-PRO-GLU-THR-GLY-GLU-GLN-ILE-GLN | | Authors: | Kekez, I, Matic, S, Tomic, S, Matkovic-Calogovic, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Binding of dipeptidyl peptidase III to the oxidative stress cell sensor Kelch-like ECH-associated protein 1 is a two-step process.
J.Biomol.Struct.Dyn., 39, 2021
|
|
4UHB
 
 | | Laboratory evolved variant R-C1 of potato epoxide hydrolase StEH1 | | Descriptor: | 1,2-ETHANEDIOL, EPOXIDE HYDROLASE, GLYCEROL | | Authors: | Nilsson, M.T.I, Carlsson, A.J, Dobritzsch, D, Widersten, M. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Laboratory Evolved Enzymes Provide Snapshots of the Development of Enantioconvergence in Enzyme-Catalyzed Epoxide Hydrolysis.
Chembiochem, 17, 2016
|
|
6TJG
 
 | | Crystal structure of the computationally designed Cake8 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cake8 | | Authors: | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
4UPF
 
 | | Assembly principles of the unique cage formed by the ATPase RavA hexamer and the lysine decarboxylase LdcI decamer | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
6TOV
 
 | | Crystal Structure of Teicoplanin Aglycone | | Descriptor: | DIMETHYL SULFOXIDE, Teicoplanin Aglycone | | Authors: | Belviso, B.D, Carrozzini, B, Caliandro, R, Altomare, C.D, Bolognino, I, Cellamare, S. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (0.767 Å) | | Cite: | Enantiomeric Separation and Molecular Modelling of Bioactive 4-Aryl-3,4-dihydropyrimidin-2(1H)-one Ester Derivatives on Teicoplanin-Based Chiral Stationary Phase
Separations, 2022
|
|
6TTA
 
 | | Haddock model of NDM-1/quercetin complex | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
6LCC
 
 | |
4UKD
 
 | | UMP/CMP KINASE FROM SLIME MOLD COMPLEXED WITH ADP, UDP, BERYLLIUM FLUORIDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM DIFLUORIDE, MAGNESIUM ION, ... | | Authors: | Schlichting, I, Reinstein, J. | | Deposit date: | 1997-05-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of active conformations of UMP kinase from Dictyostelium discoideum suggest phosphoryl transfer is associative.
Biochemistry, 36, 1997
|
|
4UEY
 
 | |
6TN7
 
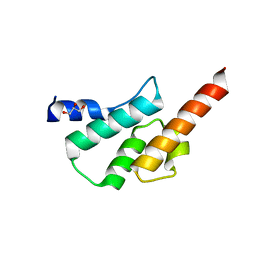 | | Crystal structure of the human Arc C-lobe | | Descriptor: | Activity-regulated cytoskeleton-associated protein, GLYCEROL | | Authors: | Hallin, E.I, Bramham, C.R, Kursula, P. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural properties and peptide ligand binding of the capsid homology domains of human Arc.
Biochem Biophys Rep, 26, 2021
|
|
6TT8
 
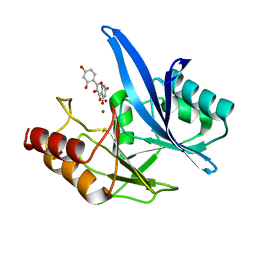 | | Haddock model of NDM-1/morin complex | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
6TTC
 
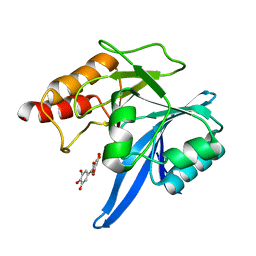 | | Haddock model of NDM-1/myricetin complex | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
4UYL
 
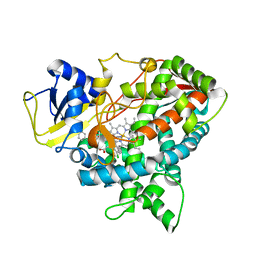 | | Crystal structure of sterol 14-alpha demethylase (CYP51B) from a pathogenic filamentous fungus Aspergillus fumigatus in complex with VNI | | Descriptor: | 14-ALPHA STEROL DEMETHYLASE, N-[(1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2014-09-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure-Functional Characterization of Cytochrome P450 Sterol Alpha-Demethylase (Cyp51B) from Aspergillus Fumigatus and Molecular Basis for the Development of Antifungal Drugs
J.Biol.Chem., 290, 2015
|
|
6LSO
 
 | |
6LSP
 
 | |
4UQM
 
 | | Crystal structure determination of uracil-DNA N-glycosylase (UNG) from Deinococcus radiodurans in complex with DNA - new insights into the role of the Leucine-loop for damage recognition and repair | | Descriptor: | 5'-D(*CP*CP*TP*AP*TP*CP*CP*AP*AAB*GP*TP*CP*TP*CP*CP*G)-3', 5'-D(*GP*CP*GP*GP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*AP*G)-3', CHLORIDE ION, ... | | Authors: | Pedersen, H.L, Johnson, K.A, McVey, C.E, Leiros, I, Moe, E. | | Deposit date: | 2014-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure determination of uracil-DNA N-glycosylase from Deinococcus radiodurans in complex with DNA.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
7OPR
 
 | | Rab27a fusion with Slp2a-RBDa1 effector covalent adduct with CB1 in C123 | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Jamshidiha, M, Tersa, M, Lanyon-Hogg, T, Perez-Dorado, I, Sutherell, C.L, De Vita, E, Morgan, R.M.L, Tate, E.W, Cota, E. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of the first structurally validated covalent ligands of the small GTPase RAB27A.
Rsc Med Chem, 13, 2022
|
|
8ASL
 
 | | RCII/PSI complex, class 2 | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, Z, Vercellino, I, Knoppova, J, Sobotka, R, Murray, J.W, Nixon, P.J, Sazanov, L.A, Komenda, J. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis.
Nat Commun, 14, 2023
|
|
6WYV
 
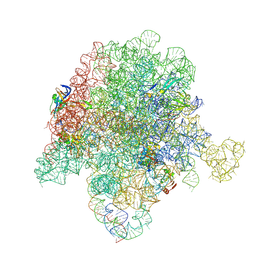 | | E. coli 50S ribosome bound to compounds 47 and VS1 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
8ASP
 
 | | RCII/PSI complex, focused refinement of PSI | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, Z, Vercellino, I, Knoppova, J, Sobotka, R, Murray, J.W, Nixon, P.J, Sazanov, L.A, Komenda, J. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis.
Nat Commun, 14, 2023
|
|
