4Y1J
 
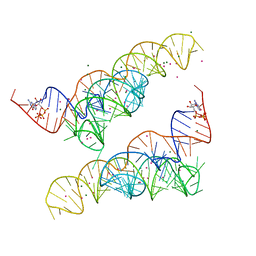 | |
4DP0
 
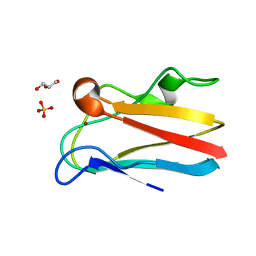 | | The 1.5 Angstrom crystal structure of oxidized (CuII) poplar plastocyanin B at pH 4.0 | | 分子名称: | COPPER (II) ION, GLYCEROL, Plastocyanin B, ... | | 著者 | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | 登録日 | 2012-02-13 | | 公開日 | 2013-02-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
6S7A
 
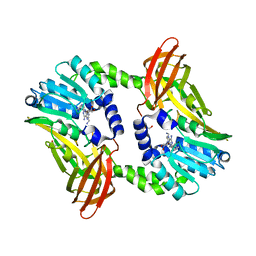 | | Crystal structure of CARM1 in complex with inhibitor AA175 | | 分子名称: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-azanylpropyl-[3-(pyridin-2-ylamino)propyl]amino]methyl]oxolane-3,4-diol, GLYCEROL, Histone-arginine methyltransferase CARM1 | | 著者 | Gunnell, E.A, Al-Noori, A, Dowden, J, Dreveny, I. | | 登録日 | 2019-07-04 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6AVN
 
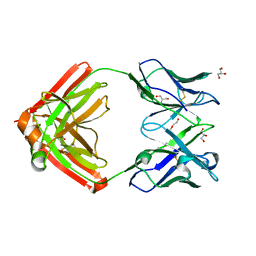 | |
3GD3
 
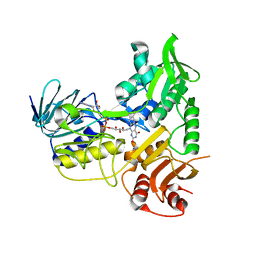 | |
2WWE
 
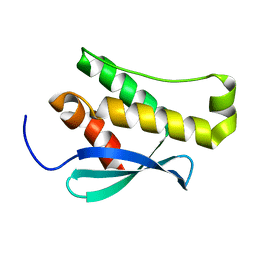 | | Crystal structure of the phox homology domain of human phosphoinositide-3-kinase-C2-gamma | | 分子名称: | PHOSPHOINOSITIDE-3-KINASE, CLASS 2, GAMMA POLYPEPTIDE | | 著者 | Roos, A.K, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotyenova, T, Kotzch, A, Kraulis, P, Markova, N, Moche, M, Nielsen, T.K, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Van Der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | 登録日 | 2009-10-22 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal Structure of the Phox Homology Domain of Human Phosphoinositide-3-Kinase-C2-Gamma
To be Published
|
|
1IFH
 
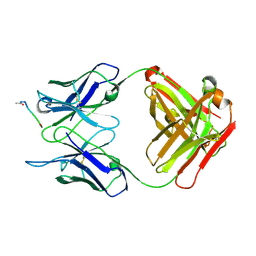 | |
4Y06
 
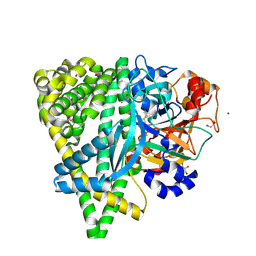 | | Crystal structure of the DAP BII (G675R) dipeptide complex | | 分子名称: | Dipeptidyl aminopeptidase BII, GLUTAMIC ACID, GLYCEROL, ... | | 著者 | Sakamoto, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2015-02-05 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
6B6I
 
 | | 2.4A resolution structure of human Norovirus GII.4 protease | | 分子名称: | 3C-like protease | | 著者 | Muzzarelli, K.M, Kuiper, B.D, Spellmon, N.S, Hackett, J, Brunzelle, J.S, Kovari, I.A, Amblard, F, Yang, Z, Schinazi, R.F, Kovari, L.C. | | 登録日 | 2017-10-02 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structural and Antiviral Studies of the Human Norovirus GII.4 Protease.
Biochemistry, 58, 2019
|
|
6EP7
 
 | | ARABIDOPSIS THALIANA GSTU23, GSH bound | | 分子名称: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase U23, ... | | 著者 | Tossounian, M.A, Van Molle, I, Wahni, K, Jacques, S, Vertommen, D, Gevaert, K, Van Breusegem, F, Young, D, Rosado, L, Messens, J. | | 登録日 | 2017-10-11 | | 公開日 | 2018-04-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.947 Å) | | 主引用文献 | Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage.
Biochim. Biophys. Acta, 1862, 2018
|
|
5MWJ
 
 | | Structure Enabled Discovery of a Stapled Peptide Inhibitor to Target the Oncogenic Transcriptional Repressor TLE1 | | 分子名称: | DIMETHYL SULFOXIDE, Transducin-like enhancer protein 1, pepide inhibtor | | 著者 | McGrath, S, Tortorici, M, Vidler, L, Drouin, L, Westwood, I, Gimeson, P, Van Montfort, R, Hoelder, S. | | 登録日 | 2017-01-18 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structure-Enabled Discovery of a Stapled Peptide Inhibitor to Target the Oncogenic Transcriptional Repressor TLE1.
Chemistry, 23, 2017
|
|
6AOP
 
 | |
6B84
 
 | |
1QO6
 
 | |
5MYF
 
 | | Convergent evolution involving dimeric and trimeric dUTPases in signalling. | | 分子名称: | dUTPase from DI S. aureus phage | | 著者 | Donderis, J, Bowring, J, Maiques, E, Ciges-Tomas, J.R, Alite, C, Mehmedov, I, Tormo-Mas, M.A, Penades, J.R, Marina, A. | | 登録日 | 2017-01-26 | | 公開日 | 2017-09-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Convergent evolution involving dimeric and trimeric dUTPases in pathogenicity island mobilization.
PLoS Pathog., 13, 2017
|
|
4JBS
 
 | | Crystal structure of the human Endoplasmic Reticulum Aminopeptidase 2 in complex with PHOSPHINIC PSEUDOTRIPEPTIDE inhibitor. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | 著者 | Saridakis, E, Birtley, J, Stratikos, E, Mavridis, I.M. | | 登録日 | 2013-02-20 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.789 Å) | | 主引用文献 | Rationally designed inhibitor targeting antigen-trimming aminopeptidases enhances antigen presentation and cytotoxic T-cell responses.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6AOS
 
 | |
6S70
 
 | | Crystal structure of CARM1 in complex with inhibitor UM251 | | 分子名称: | 1-[5-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]pentyl]guanidine, GLYCEROL, Histone-arginine methyltransferase CARM1 | | 著者 | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | 登録日 | 2019-07-04 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
5MTZ
 
 | | Crystal structure of a long form RNase Z from yeast | | 分子名称: | PHOSPHATE ION, Ribonuclease Z, ZINC ION | | 著者 | Li de la Sierra-Gallay, I, Miao, M, van Tilbeurgh, H. | | 登録日 | 2017-01-11 | | 公開日 | 2017-06-21 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | The crystal structure of Trz1, the long form RNase Z from yeast.
Nucleic Acids Res., 45, 2017
|
|
6S79
 
 | | Crystal structure of CARM1 in complex with inhibitor AA183 | | 分子名称: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-(pyridin-2-ylamino)propyl]amino]-2-azanyl-butanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1 | | 著者 | Gunnell, E.A, Al-Noori, A, Dowden, J, Dreveny, I. | | 登録日 | 2019-07-04 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
5MUI
 
 | | Glycoside hydrolase BT_0996 | | 分子名称: | Beta-galactosidase, beta-L-arabinofuranose-(1-2)-alpha-L-rhamnopyranose-(1-2)-[alpha-L-rhamnopyranose-(1-3)]alpha-L-arabinopyranose-(1-4)-[4-O-[(1R)-1-hydroxyethyl]-2-O-methyl-alpha-L-fucopyranose-(1-2)]beta-D-galactopyranose-(1-2)-alpha-D-aceric acid-(1-3)-alpha-L-rhamnopyranose | | 著者 | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | 登録日 | 2017-01-13 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
4NWB
 
 | | Crystal structure of Mrt4 | | 分子名称: | SULFATE ION, mRNA turnover protein 4 | | 著者 | Holdermann, I, Sinning, I. | | 登録日 | 2013-12-06 | | 公開日 | 2014-03-26 | | 最終更新日 | 2014-04-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
3G51
 
 | |
5MSI
 
 | |
1QWM
 
 | | Structure of Helicobacter pylori catalase with formic acid bound | | 分子名称: | AZIDE ION, FORMIC ACID, KatA catalase, ... | | 著者 | Loewen, P.C, Carpena, X, Perez-Luque, R, Rovira, C, Haas, R, Odenbreit, S, Nicholls, P, Fita, I. | | 登録日 | 2003-09-02 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of Helicobacter pylori Catalase, with and without Formic Acid Bound, at 1.6 A Resolution
Biochemistry, 43, 2004
|
|
