1NQW
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 5-(6-D-ribitylamino-2,4(1H,3H)pyrimidinedione-5-yl)-1-pentyl-phosphonic acid | | 分子名称: | 5-(6-D-RIBITYLAMINO-2,4(1H,3H)PYRIMIDINEDIONE-5-YL) PENTYL-1-PHOSPHONIC ACID, 6,7-dimethyl-8-ribityllumazine synthase | | 著者 | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | 登録日 | 2003-01-23 | | 公開日 | 2004-01-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
6T4D
 
 | | Crystal structure of Plasmodium falciparum Morn1 | | 分子名称: | Morn1, ZINC ION | | 著者 | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | 登録日 | 2019-10-13 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
1NQX
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 3-(7-hydroxy-8-ribityllumazine-6-yl)propionic acid | | 分子名称: | 3-(7-HYDROXY-8-RIBITYLLUMAZINE-6-YL) PROPIONIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | 著者 | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | 登録日 | 2003-01-23 | | 公開日 | 2004-01-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1BIX
 
 | | THE CRYSTAL STRUCTURE OF THE HUMAN DNA REPAIR ENDONUCLEASE HAP1 SUGGESTS THE RECOGNITION OF EXTRA-HELICAL DEOXYRIBOSE AT DNA ABASIC SITES | | 分子名称: | AP ENDONUCLEASE 1, PLATINUM (II) ION, SAMARIUM (III) ION | | 著者 | Gorman, M.A, Morera, S, Rothwell, D.G, De La Fortelle, E, Mol, C.D, Tainer, J.A, Hickson, I.D, Freemont, P.S. | | 登録日 | 1998-06-19 | | 公開日 | 1999-06-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites.
EMBO J., 16, 1997
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | 分子名称: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | 著者 | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | 登録日 | 2020-10-27 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
1ISW
 
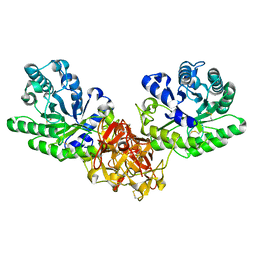 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylobiose | | 分子名称: | beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-D-xylanase | | 著者 | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | 登録日 | 2001-12-27 | | 公開日 | 2002-02-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
6T7O
 
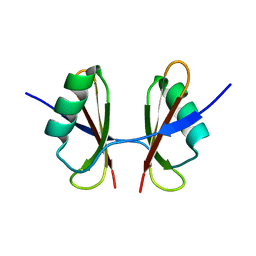 | | X-ray structure of the C-terminal domain of S. aureus Hibernating Promoting Factor (CTD-SaHPF) | | 分子名称: | Ribosome hibernation promotion factor | | 著者 | Fatkhullin, B.F, Khusainov, I.S, Ayupov, R.K, Gabdulkhakov, A.G, Tishchenko, S.V, Validov, S.Z, Yusupov, M.M. | | 登録日 | 2019-10-22 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.60003626 Å) | | 主引用文献 | Dimerization of long hibernation promoting factor from Staphylococcus aureus: Structural analysis and biochemical characterization.
J.Struct.Biol., 209, 2020
|
|
6TA6
 
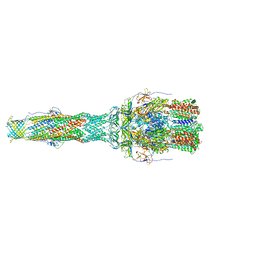 | | MexAB assembly of the Pseudomonas MexAB-OprM efflux pump reconstituted in nanodiscs | | 分子名称: | Efflux pump membrane transporter, MexA family multidrug efflux RND transporter periplasmic adaptor subunit, Outer membrane protein OprM | | 著者 | Glavier, M, Schoehn, G, Taveau, J.C, Phan, G, Daury, L, Lambert, O, Broutin, I. | | 登録日 | 2019-10-29 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex.
Nat Commun, 11, 2020
|
|
7Z04
 
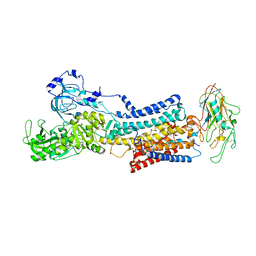 | | 10 mM Rb+ soak of beryllium fluoride inhibited Na+,K+-ATPase, E2-BeFx (rigid body model) | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, FXYD domain-containing ion transport regulator, MAGNESIUM ION, ... | | 著者 | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shasavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | 登録日 | 2022-02-22 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (7.5 Å) | | 主引用文献 | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
7YZR
 
 | | 50 mM Rb+ soak of beryllium fluoride inhibited Na+,K+-ATPase, E2-BeFx (rigid body model) | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, FXYD domain-containing ion transport regulator, MAGNESIUM ION, ... | | 著者 | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shasavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | 登録日 | 2022-02-21 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (6.92 Å) | | 主引用文献 | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
6T69
 
 | | Crystal structure of Toxoplasma gondii Morn1(V shape) | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Membrane occupation and recognition nexus protein MORN1, ... | | 著者 | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | 登録日 | 2019-10-18 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
1PW2
 
 | | APO STRUCTURE OF HUMAN CYCLIN-DEPENDENT KINASE 2 | | 分子名称: | Cell division protein kinase 2 | | 著者 | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | 登録日 | 2003-06-30 | | 公開日 | 2003-12-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the
activation loop.
Structure, 11, 2003
|
|
7KSB
 
 | | Crystal structure on Act c 10.0101 | | 分子名称: | Non-specific lipid-transfer protein 1, SULFATE ION | | 著者 | Pote, S, O'Malley, A, Gawlicka-Chruszcz, A, Giangrieco, I, Ciardiello, M.A, Chruszcz, M. | | 登録日 | 2020-11-21 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural Characterization of Act c 10.0101 and Pun g 1.0101-Allergens from the Non-Specific Lipid Transfer Protein Family.
Molecules, 26, 2021
|
|
1SQN
 
 | | Progesterone Receptor Ligand Binding Domain with bound Norethindrone | | 分子名称: | (14beta,17alpha)-17-ethynyl-17-hydroxyestr-4-en-3-one, progesterone receptor | | 著者 | Williams, S.P, Madauss, K.P, Deng, J.-S, Austin, R.J.H, Lambert, M.H, McLay, I, Pritchard, J, Short, S.A, Stewart, E.L, Uings, I.J. | | 登録日 | 2004-03-19 | | 公開日 | 2004-07-27 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | Progesterone receptor ligand binding pocket flexibility: crystal structures of the norethindrone and mometasone furoate complexes
J.Med.Chem., 47, 2004
|
|
7KVS
 
 | | Human CYP3A4 bound to an inhibitor | | 分子名称: | 1,2-ETHANEDIOL, Cytochrome P450 3A4, GLYCEROL, ... | | 著者 | Sevrioukova, I. | | 登録日 | 2020-11-28 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Rational Design of CYP3A4 Inhibitors: A One-Atom Linker Elongation in Ritonavir-Like Compounds Leads to a Marked Improvement in the Binding Strength.
Int J Mol Sci, 22, 2021
|
|
2C26
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | 著者 | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | 登録日 | 2005-09-26 | | 公開日 | 2005-10-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
6T8H
 
 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | 分子名称: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | 著者 | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | 登録日 | 2019-10-24 | | 公開日 | 2020-03-04 | | 最終更新日 | 2020-04-08 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
1EG2
 
 | | CRYSTAL STRUCTURE OF RHODOBACTER SPHEROIDES (N6 ADENOSINE) METHYLTRANSFERASE (M.RSRI) | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MODIFICATION METHYLASE RSRI | | 著者 | Scavetta, R.D, Thomas, C.B, Walsh, M.A, Szegedi, S, Joachimiak, A, Gumport, R.I, Churchill, M.E.A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2000-02-11 | | 公開日 | 2000-10-18 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases.
Nucleic Acids Res., 28, 2000
|
|
1J0B
 
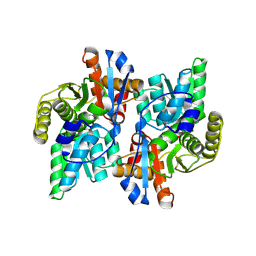 | | Crystal Structure Analysis of the ACC deaminase homologue complexed with inhibitor | | 分子名称: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | 著者 | Fujino, A, Ose, T, Honma, M, Yao, M, Tanaka, I. | | 登録日 | 2002-11-12 | | 公開日 | 2003-05-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and enzymatic properties of 1-aminocyclopropane-1-carboxylate deaminase homologue from Pyrococcus horikoshii
J.Mol.Biol., 341, 2004
|
|
1MU2
 
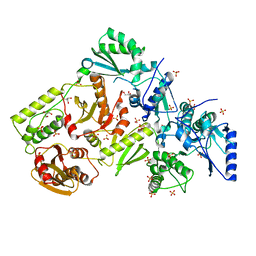 | | CRYSTAL STRUCTURE OF HIV-2 REVERSE TRANSCRIPTASE | | 分子名称: | GLYCEROL, HIV-2 RT, SULFATE ION | | 著者 | Ren, J, Bird, L.E, Chamberlain, P.P, Stewart-Jones, G.B, Stuart, D.I, Stammers, D.K. | | 登録日 | 2002-09-23 | | 公開日 | 2002-10-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to non-nucleoside inhibitors
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6CMZ
 
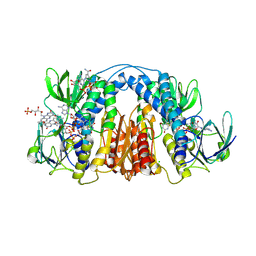 | | 2.3 Angstrom Resolution Crystal Structure of Dihydrolipoamide Dehydrogenase from Burkholderia cenocepacia in Complex with FAD and NAD | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, D-MALATE, ... | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-03-06 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | 2.3 Angstrom Resolution Crystal Structure of Dihydrolipoamide Dehydrogenase from Burkholderia cenocepacia in Complex with FAD and NAD.
To Be Published
|
|
5TTW
 
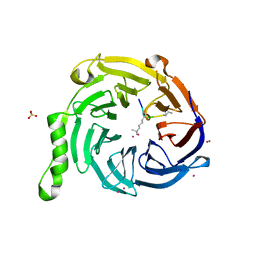 | | Crystal Structure of EED in Complex with UNC4859 | | 分子名称: | Polycomb protein EED, SULFATE ION, UNC4859, ... | | 著者 | The, J, Barnash, K.D, Brown, P.J, Edwards, A.M, Bountra, C, Frye, S.V, James, L.I, Arrowsmith, C.H. | | 登録日 | 2016-11-04 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Discovery of Peptidomimetic Ligands of EED as Allosteric Inhibitors of PRC2.
ACS Comb Sci, 19, 2017
|
|
7KVQ
 
 | | Human CYP3A4 bound to an inhibitor | | 分子名称: | 1,2-ETHANEDIOL, Cytochrome P450 3A4, GLYCEROL, ... | | 著者 | Sevrioukova, I. | | 登録日 | 2020-11-28 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Rational Design of CYP3A4 Inhibitors: A One-Atom Linker Elongation in Ritonavir-Like Compounds Leads to a Marked Improvement in the Binding Strength.
Int J Mol Sci, 22, 2021
|
|
1N5N
 
 | | Crystal Structure of Peptide Deformylase from Pseudomonas aeruginosa | | 分子名称: | GLYCEROL, Peptide deformylase, ZINC ION | | 著者 | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | 登録日 | 2002-11-06 | | 公開日 | 2003-06-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1IE6
 
 | | SOLUTION STRUCTURE OF IMPERATOXIN A | | 分子名称: | IMPERATOXIN A | | 著者 | Lee, C.W, Takeuchi, K, Takahashi, H, Sato, K, Shimada, I, Kim, D.H, Kim, J.I. | | 登録日 | 2001-04-07 | | 公開日 | 2003-06-10 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular basis of the high-affinity activation of type 1 ryanodine receptors by imperatoxin A.
Biochem.J., 377, 2004
|
|
