7A76
 
 | |
7RLU
 
 | | Structure of ALDH1L1 (10-formyltetrahydrofolate dehydrogenase) in complex with NADP | | 分子名称: | 4'-PHOSPHOPANTETHEINE, Cytosolic 10-formyltetrahydrofolate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Tsybovsky, Y, Sereda, V, Golczak, M, Krupenko, N.I, Krupenko, S.A. | | 登録日 | 2021-07-26 | | 公開日 | 2022-01-12 | | 最終更新日 | 2022-02-02 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure of putative tumor suppressor ALDH1L1.
Commun Biol, 5, 2022
|
|
5NOP
 
 | | Structure of Mojiang virus attachment glycoprotein | | 分子名称: | Attachment glycoprotein, CHLORIDE ION | | 著者 | Rissanen, I.R, Ahmed, A.A, Beaty, S, Azarm, K, Hong, P, Nambulli, S, Duprex, P.W, Lee, B, Bowden, T.A. | | 登録日 | 2017-04-12 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Idiosyncratic Mojiang virus attachment glycoprotein directs a host-cell entry pathway distinct from genetically related henipaviruses.
Nat Commun, 8, 2017
|
|
7RLZ
 
 | |
7RLW
 
 | |
7RLV
 
 | |
7RM0
 
 | |
7RLY
 
 | |
7RM3
 
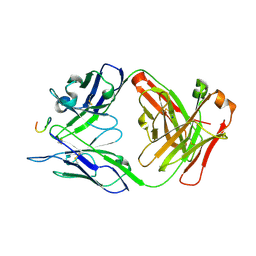 | | Antibody 2E10.E9 in complex with P. vivax CSP peptide ANGAGNQPGANGAGNQPGANGAGGQAA | | 分子名称: | 2E10.E9 Fab heavy chain, 2E10.E9 Fab light chain, ACETATE ION, ... | | 著者 | Kucharska, I, Ivanochko, D, Julien, J.P. | | 登録日 | 2021-07-26 | | 公開日 | 2022-01-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural basis of Plasmodium vivax inhibition by antibodies binding to the circumsporozoite protein repeats.
Elife, 11, 2022
|
|
7A5V
 
 | | CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | 登録日 | 2020-08-22 | | 公開日 | 2020-11-18 | | 最終更新日 | 2020-11-25 | | 実験手法 | ELECTRON MICROSCOPY (1.7 Å) | | 主引用文献 | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
5NDW
 
 | | Crystal structure of aminoglycoside TC007 bound to the yeast 80S ribosome | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | 登録日 | 2017-03-09 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6ZMP
 
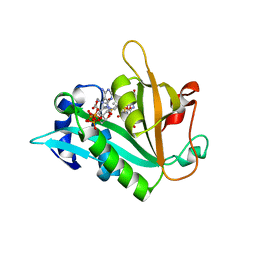 | |
7R2V
 
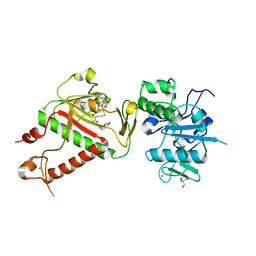 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | 著者 | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | 登録日 | 2022-02-06 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
1E8A
 
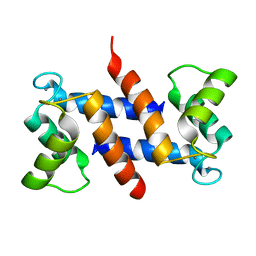 | | The three-dimensional structure of human S100A12 | | 分子名称: | CALCIUM ION, S100A12 | | 著者 | Moroz, O.V, Antson, A.A, Murshudov, G.N, Maitland, N.J, Dodson, G.G, Wilson, K.S, Skibshoj, I, Lukanidin, E.M, Bronstein, I.B. | | 登録日 | 2000-09-18 | | 公開日 | 2001-01-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The Three-Dimensional Structure of Human S100A12
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7A5Y
 
 | | Crystal structure of tetrameric human H215A-SAMHD1 (residues 109-626) with Rp-dGTP-alphaS (T8T) and Mg | | 分子名称: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | 著者 | Morris, E.R, Kunzelmann, S, Caswell, S.J, Purkiss, A, Taylor, I.A. | | 登録日 | 2020-08-24 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Probing the Catalytic Mechanism and Inhibition of SAMHD1 Using the Differential Properties of R p - and S p -dNTP alpha S Diastereomers.
Biochemistry, 60, 2021
|
|
5OBM
 
 | | Crystal structure of Gentamicin bound to the yeast 80S ribosome | | 分子名称: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 18S ribosomal RNA, 25S ribosomal RNA, ... | | 著者 | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | 登録日 | 2017-06-28 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6ZI3
 
 | | Crystal structure of OleP-6DEB bound to L-rhamnose | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, ... | | 著者 | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | 登録日 | 2020-06-24 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
2XFX
 
 | | cattle MHC class I N01301 presenting an 11mer from Theileria parva | | 分子名称: | BETA-2-MICROGLOBULIN, MHC CLASS 1, UNCHARACTERIZED PROTEIN | | 著者 | Macdonald, I.K, Harkiolaki, M, Hunt, L, Morrison, W.I, Connelley, T, Graham, S.P, Jones, E.Y, Flower, D.R, Ellis, S.A. | | 登録日 | 2010-05-28 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Mhc Class I Bound to an Immunodominant Theileria Parva Epitope Demonstrates Unconventional Presentation to T Cell Receptors.
Plos Pathog., 6, 2010
|
|
1SJM
 
 | | Nitrite bound copper containing nitrite reductase | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, COPPER (II) ION, ... | | 著者 | Tocheva, E.I, Rosell, F.I, Mauk, A.G, Murphy, M.E.P. | | 登録日 | 2004-03-03 | | 公開日 | 2004-06-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Side-on copper-nitrosyl coordination by nitrite reductase.
Science, 304, 2004
|
|
1CD1
 
 | |
6ZDG
 
 | | Association of three complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-14 | | 公開日 | 2020-07-29 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZCZ
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in ternary complex with EY6A Fab and a nanobody. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EY6A heavy chain, ... | | 著者 | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-12 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZER
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with EY6A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | 著者 | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-16 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6BZ2
 
 | | Crystal structure of wild-type HIV-1 protease with a novel HIV-1 inhibitor GRL-14213A of 6-5-5-ring fused crown-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2017-12-22 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Design of Highly Potent, Dual-Acting and Central-Nervous-System-Penetrating HIV-1 Protease Inhibitors with Excellent Potency against Multidrug-Resistant HIV-1 Variants.
ChemMedChem, 13, 2018
|
|
7R5Y
 
 | |
