1QA1
 
 | | TAILSPIKE PROTEIN, MUTANT V331G | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Baxa, U, Steinbacher, S, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-04-10 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
1QQ1
 
 | | TAILSPIKE PROTEIN, MUTANT E359G | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Schuler, B, Furst, F, Osterroth, F, Steinbacher, S, Huber, R, Seckler, R. | | Deposit date: | 1999-06-10 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Plasticity and steric strain in a parallel beta-helix: rational mutations in the P22 tailspike protein.
Proteins, 39, 2000
|
|
1QA3
 
 | | TAILSPIKE PROTEIN, MUTANT A334I | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Baxa, U, Steinbacher, S, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-04-10 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
1ORW
 
 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Peptidomimetic Inhibitor | | Descriptor: | (2S)-PYRROLIDIN-2-YLMETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2H6T
 
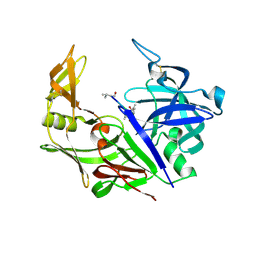 | | Secreted aspartic proteinase (Sap) 3 from Candida albicans complexed with pepstatin A | | Descriptor: | Candidapepsin-3, ZINC ION, pepstatin A | | Authors: | Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2006-06-01 | | Release date: | 2007-06-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the secreted aspartic proteinase 3 from Candida albicans and its complex with pepstatin A.
Proteins, 68, 2007
|
|
2H6S
 
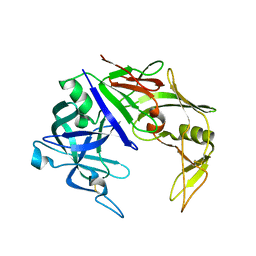 | | Secreted aspartic proteinase (Sap) 3 from Candida albicans | | Descriptor: | Candidapepsin-3, ZINC ION | | Authors: | Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2006-06-01 | | Release date: | 2007-06-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the secreted aspartic proteinase 3 from Candida albicans and its complex with pepstatin A.
Proteins, 68, 2007
|
|
1REI
 
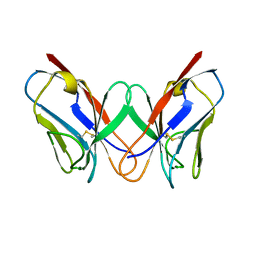 | | THE MOLECULAR STRUCTURE OF A DIMER COMPOSED OF THE VARIABLE PORTIONS OF THE BENCE-JONES PROTEIN REI REFINED AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | BENCE-JONES PROTEIN REI (LIGHT CHAIN) | | Authors: | Epp, O, Lattman, E.E, Colman, P, Fehlhammer, H, Bode, W, Schiffer, M, Huber, R, Palm, W. | | Deposit date: | 1976-03-17 | | Release date: | 1976-05-19 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular structure of a dimer composed of the variable portions of the Bence-Jones protein REI refined at 2.0-A resolution.
Biochemistry, 14, 1975
|
|
1ORV
 
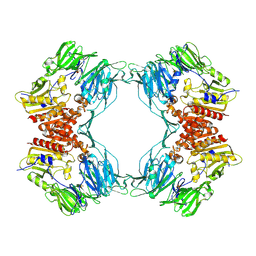 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1SEZ
 
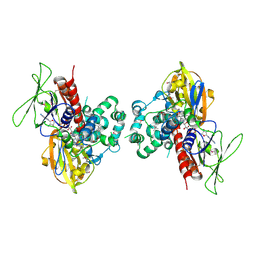 | | Crystal Structure of Protoporphyrinogen IX Oxidase | | Descriptor: | 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, 4-BROMO-3-(5'-CARBOXY-4'-CHLORO-2'-FLUOROPHENYL)-1-METHYL-5-TRIFLUOROMETHYL-PYRAZOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Koch, M, Breithaupt, C, Kiefersauer, R, Freigang, J, Huber, R, Messerschmidt, A. | | Deposit date: | 2004-02-19 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis.
Embo J., 23, 2004
|
|
1TOC
 
 | |
2F9P
 
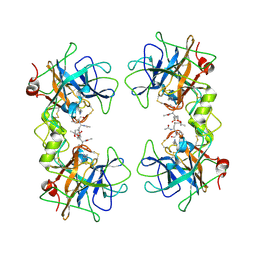 | | Crystal Structure of the Recombinant Human Alpha I Tryptase Mutant D216G in Complex with Leupeptin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Leupeptin, ... | | Authors: | Rohr, K.B, Selwood, T, Marquardt, U, Huber, R, Schechter, N.M, Bode, W, Than, M.E. | | Deposit date: | 2005-12-06 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Structures of Free and Leupeptin-complexed Human alpha I-Tryptase Mutants: Indication for an alpha to beta-Tryptase Transition
J.Mol.Biol., 357, 2005
|
|
2F9N
 
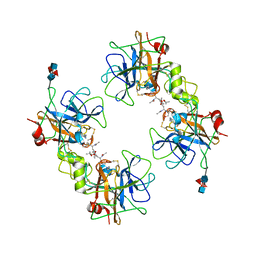 | | Crystal Structure of the Recombinant Human Alpha I Tryptase Mutant K192Q/D216G in Complex with Leupeptin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Leupeptin, ... | | Authors: | Rohr, K.B, Selwood, T, Marquardt, U, Huber, R, Schechter, N.M, Bode, W, Than, M.E. | | Deposit date: | 2005-12-06 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray Structures of Free and Leupeptin-complexed Human alpha I-Tryptase Mutants: Indication for an alpha to beta-Tryptase Transition
J.Mol.Biol., 357, 2005
|
|
1SVG
 
 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 4 | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ALPHA FORM, N-{(3R,4R)-4-[4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-BENZOYLAMINO]-AZEPAN-3-YL}ISONICOTINAMIDE, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.-G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
1QA2
 
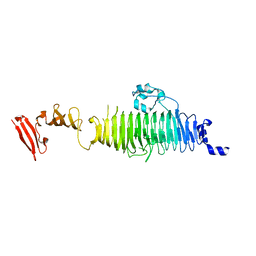 | | TAILSPIKE PROTEIN, MUTANT A334V | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Baxa, U, Steinbacher, S, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-04-10 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
1JJ9
 
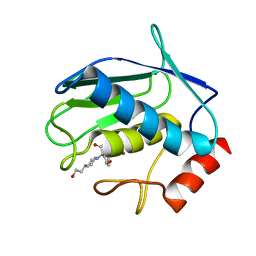 | | Crystal Structure of MMP8-Barbiturate Complex Reveals Mechanism for Collagen Substrate Recognition | | Descriptor: | 2-HYDROXY-5-[4-(2-HYDROXY-ETHYL)-PIPERIDIN-1-YL]-5-PHENYL-1H-PYRIMIDINE-4,6-DIONE, CALCIUM ION, Matrix Metalloproteinase 8, ... | | Authors: | Brandstetter, H, Grams, F, Glitz, D, Lang, A, Huber, R, Bode, W, Krell, H.-W, Engh, R.A. | | Deposit date: | 2001-07-04 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 1.8-A crystal structure of a matrix metalloproteinase 8-barbiturate inhibitor complex reveals a previously unobserved mechanism for collagenase substrate recognition.
J.Biol.Chem., 276, 2001
|
|
1SVE
 
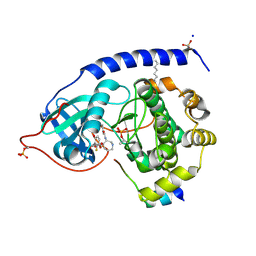 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 1 | | Descriptor: | (4R)-4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-BENZOIC ACID (3R)-3-[(PYRIDINE-4-CARBONYL)AMINO]-AZEPAN-4-YL ESTER, N-OCTANOYL-N-METHYLGLUCAMINE, SODIUM ION, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.-G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
2F9O
 
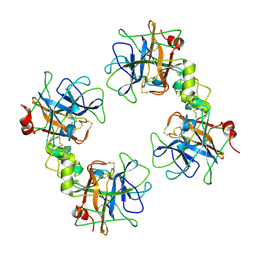 | | Crystal Structure of the Recombinant Human Alpha I Tryptase Mutant D216G | | Descriptor: | Tryptase alpha-1, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Rohr, K.B, Selwood, T, Marquardt, U, Huber, R, Schechter, N.M, Bode, W, Than, M.E. | | Deposit date: | 2005-12-06 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of Free and Leupeptin-complexed Human alpha I-Tryptase Mutants: Indication for an alpha to beta-Tryptase Transition
J.Mol.Biol., 357, 2005
|
|
1TLE
 
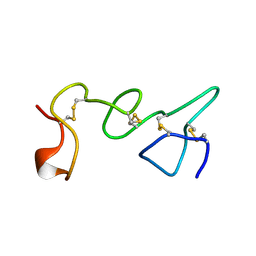 | | LE (LAMININ-TYPE EGF-LIKE) MODULE GIII4 IN SOLUTION AT PH 3.5 AND 290 K, NMR, 14 STRUCTURES | | Descriptor: | LAMININ | | Authors: | Baumgartner, R, Czisch, M, Mayer, U, Schl, E.P, Huber, R, Timpl, R, Holak, T.A. | | Deposit date: | 1996-01-26 | | Release date: | 1997-02-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the nidogen binding LE module of the laminin gamma1 chain in solution.
J.Mol.Biol., 257, 1996
|
|
1SVH
 
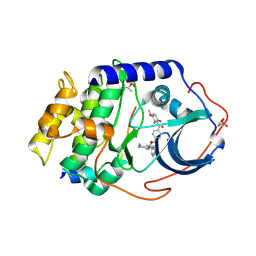 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 8 | | Descriptor: | (3R,4S)-N-(4-{TRANS-2-[4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-PHENYL]-VINYL}-AZEPAN-3-YL)-ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
1CTS
 
 | |
1WQJ
 
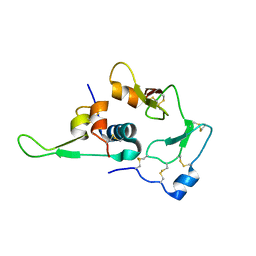 | | Structural Basis for the Regulation of Insulin-Like Growth Factors (IGFs) by IGF Binding Proteins (IGFBPs) | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor binding protein 4 | | Authors: | Siwanowicz, I, Popowicz, G.M, Wisniewska, M, Huber, R, Kuenkele, K.P, Lang, K, Engh, R.A, Holak, T.A. | | Deposit date: | 2004-09-29 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the regulation of insulin-like growth factors by IGF binding proteins
Structure, 13, 2005
|
|
1WYX
 
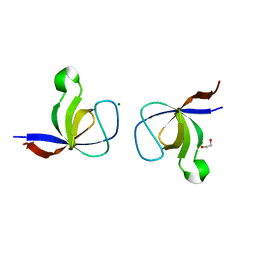 | | The Crystal Structure of the p130Cas SH3 Domain at 1.1 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CRK-associated substrate, MAGNESIUM ION | | Authors: | Wisniewska, M, Bossenmaier, B, Georges, G, Hesse, F, Dangl, M, Kuenkele, K.P, Ioannidis, I, Huber, R, Engh, R.A. | | Deposit date: | 2005-02-17 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The 1.1 A resolution crystal structure of the p130cas SH3 domain and ramifications for ligand selectivity
J.Mol.Biol., 347, 2005
|
|
1DX5
 
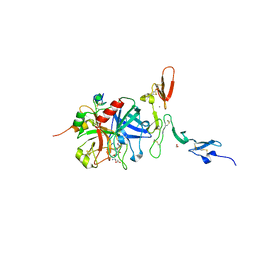 | | Crystal structure of the thrombin-thrombomodulin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FORMIC ACID, ... | | Authors: | Fuentes-Prior, P, Iwanaga, Y, Huber, R, Pagila, R, Rumennik, G, Seto, M, Morser, J, Light, D.R, Bode, W. | | Deposit date: | 1999-12-20 | | Release date: | 2000-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Anticoagulant Activity of the Thrombin-Thrombomodulin Complex
Nature, 404, 2000
|
|
1XRN
 
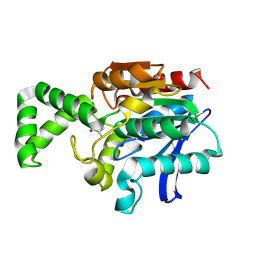 | | Crystal structure of active site F1-mutant E213Q soaked with peptide Phe-Ala | | Descriptor: | ALANINE, Proline iminopeptidase | | Authors: | Goettig, P, Brandstetter, H, Groll, M, Goehring, W, Konarev, P.V, Svergun, D.I, Huber, R, Kim, J.-S. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray snapshots of peptide processing in mutants of tricorn-interacting factor F1 from Thermoplasma acidophilum
J.Biol.Chem., 280, 2005
|
|
1CLW
 
 | | TAILSPIKE PROTEIN FROM PHAGE P22, V331A MUTANT | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S, Baxa, U, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-05-04 | | Release date: | 1999-11-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
