8F5Y
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Chen, T. | | Deposit date: | 2022-11-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A bromodomain-independent mechanism of gene regulation by the BET inhibitor JQ1: direct activation of nuclear receptor PXR.
Nucleic Acids Res., 52, 2024
|
|
8EQZ
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-C6 | | Descriptor: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-hexylbenzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | Deposit date: | 2022-10-11 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8E3N
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with rifamycin S | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, Rifamycin S | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Chen, T. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FPE
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-BP | | Descriptor: | N-[([1,1'-biphenyl]-4-yl)methyl]-N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | Deposit date: | 2023-01-04 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SZV
 
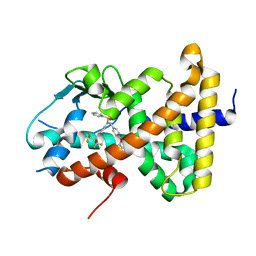 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog SJPYT-318 | | Descriptor: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-[(4-phenoxyphenyl)methyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Miller, D.J, Li, Y, Chen, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand flexibility and binding pocket malleability cooperate to allow selective PXR activation by analogs of a promiscuous nuclear receptor ligand.
Structure, 31, 2023
|
|
9BEQ
 
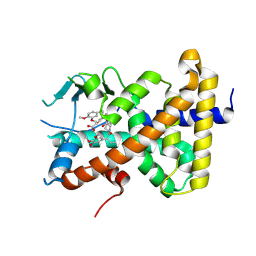 | | Crystal structure of pregnane X receptor ligand binding domain complexed with AP1867 | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, {3-[3-(3,4-DIMETHOXY-PHENYL)-1-(1-{1-[2-(3,4,5-TRIMETHOXY-PHENYL)-BUTYRYL]-PIPERIDIN-2YL}-VINYLOXY)-PROPYL]-PHENOXY}-ACETIC ACID | | Authors: | Huber, A.D, Poudel, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-04-16 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PROTAC-mediated activation, rather than degradation, of a nuclear receptor reveals complex ligand-receptor interaction network
Structure, 2024
|
|
3BCT
 
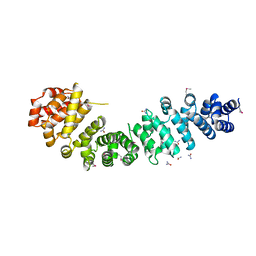 | | THE ARMADILLO REPEAT REGION FROM MURINE BETA-CATENIN | | Descriptor: | BETA-CATENIN, CHLORIDE ION, UREA | | Authors: | Huber, A.H, Nelson, W.J, Weis, W.I. | | Deposit date: | 1997-07-31 | | Release date: | 1997-11-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the armadillo repeat region of beta-catenin.
Cell(Cambridge,Mass.), 90, 1997
|
|
1CFB
 
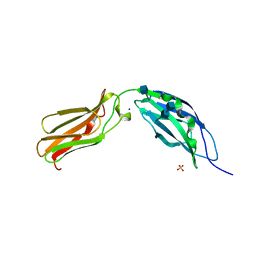 | | CRYSTAL STRUCTURE OF TANDEM TYPE III FIBRONECTIN DOMAINS FROM DROSOPHILA NEUROGLIAN AT 2.0 ANGSTROMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DROSOPHILA NEUROGLIAN, ... | | Authors: | Huber, A.H, Wang, Y.E, Bieber, A.J, Bjorkman, P.J. | | Deposit date: | 1994-08-27 | | Release date: | 1994-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of tandem type III fibronectin domains from Drosophila neuroglian at 2.0 A.
Neuron, 12, 1994
|
|
1I7W
 
 | | BETA-CATENIN/PHOSPHORYLATED E-CADHERIN COMPLEX | | Descriptor: | BETA-CATENIN, CHLORIDE ION, EPITHELIAL-CADHERIN, ... | | Authors: | Huber, A.H, Weis, W.I. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I7X
 
 | | BETA-CATENIN/E-CADHERIN COMPLEX | | Descriptor: | BETA-CATENIN, EPITHELIAL-CADHERIN | | Authors: | Huber, A.H, Weis, W.I. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
2BCT
 
 | |
6W9R
 
 | |
6W9S
 
 | |
6W9O
 
 | |
8T48
 
 | |
6EQI
 
 | | Structure of PINK1 bound to ubiquitin | | Descriptor: | GLYCEROL, Nb696, Serine/threonine-protein kinase PINK1, ... | | Authors: | Schubert, A.F, Gladkova, C, Pardon, E, Wagstaff, J.L, Freund, S.M.V, Steyaert, J, Maslen, S, Komander, D. | | Deposit date: | 2017-10-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of PINK1 in complex with its substrate ubiquitin.
Nature, 552, 2017
|
|
8SVO
 
 | | Crystal structure of pregnane X receptor ligand binding domain in complex with SJPYT-310 | | Descriptor: | (1P)-N-(5-tert-butyl-2-{[(2S)-pentan-2-yl]oxy}phenyl)-1-(2-methoxy-5-methylphenyl)-5-methyl-1H-1,2,3-triazole-4-carboxamide, DIMETHYL SULFOXIDE, Pregnane X receptor ligand binding domain fused to SRC-1 coactivator peptide | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVN
 
 | | Crystal structure of the apo form of pregnane X receptor ligand binding domain | | Descriptor: | Pregnane X receptor ligand binding domain fused to SRC-1 coactivator peptide | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVU
 
 | | Crystal structure of the L428V mutant of pregnane X receptor ligand binding domain in apo form | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1 fusion protein,Nuclear receptor coactivator 1 | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVX
 
 | | Crystal structure of the L428V mutant of pregnane X receptor ligand binding domain in complex with SJPYT-331 | | Descriptor: | DIMETHYL SULFOXIDE, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1 fusion protein,Nuclear receptor coactivator 1, ... | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
7BAE
 
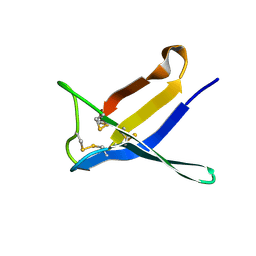 | | Crystal structure of PAFB | | Descriptor: | Antifungal protein | | Authors: | Guagnini, F, Huber, A, Alex, J.M, Marx, F, Crowley, P.B. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Porous assembly of an antifungal protein mediated by zinc and sulfonato-calix[8]arene.
J.Struct.Biol., 213, 2021
|
|
7BAD
 
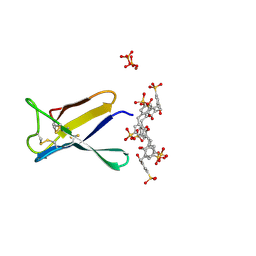 | | Crystal structure of PAFB in complex with p-sulfonato-calix[8]arene and zinc | | Descriptor: | Antifungal protein, PHOSPHATE ION, ZINC ION, ... | | Authors: | Guagnini, F, Huber, A, Alex, J.M, Marx, F, Crowley, P.B. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Porous assembly of an antifungal protein mediated by zinc and sulfonato-calix[8]arene.
J.Struct.Biol., 213, 2021
|
|
7BAF
 
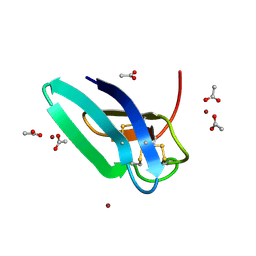 | | Crystal structure of PAFB in complex with zinc | | Descriptor: | ACETATE ION, Antifungal protein, ZINC ION | | Authors: | Guagnini, F, Huber, A, Alex, J.M, Marx, F, Crowley, P.B. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.123 Å) | | Cite: | Porous assembly of an antifungal protein mediated by zinc and sulfonato-calix[8]arene.
J.Struct.Biol., 213, 2021
|
|
8SVP
 
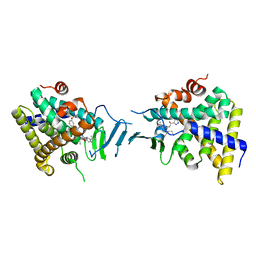 | | Crystal structure of pregnane X receptor ligand binding domain in complex with SJPYT-278 | | Descriptor: | (1P)-N-(5-tert-butyl-2-{[(3R)-hexan-3-yl]oxy}phenyl)-1-(2,5-dimethoxyphenyl)-5-methyl-1H-1,2,3-triazole-4-carboxamide, (1P)-N-(5-tert-butyl-2-{[(3S)-hexan-3-yl]oxy}phenyl)-1-(2,5-dimethoxyphenyl)-5-methyl-1H-1,2,3-triazole-4-carboxamide, Pregnane X receptor ligand binding domain fused to SRC-1 coactivator peptide | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVR
 
 | | Crystal structure of pregnane X receptor ligand binding domain in complex with SJPYT-326 | | Descriptor: | (1P)-N-(5-tert-butyl-2-{[(3S)-hexan-3-yl]oxy}phenyl)-5-methyl-1-(2,4,5-trimethoxyphenyl)-1H-1,2,3-triazole-4-carboxamide, DIMETHYL SULFOXIDE, Pregnane X receptor ligand binding domain fused to SRC-1 coactivator peptide | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
