6A6L
 
 | |
1PW4
 
 | | Crystal Structure of the Glycerol-3-Phosphate Transporter from E.Coli | | Descriptor: | Glycerol-3-phosphate transporter | | Authors: | Huang, Y, Lemieux, M.J, Song, J, Auer, M, Wang, D.N. | | Deposit date: | 2003-06-30 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and Mechanism of the Glycerol-3-Phosphate Transporter from Escherichia Coli
Science, 301, 2003
|
|
5AYW
 
 | | Structure of a membrane complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Huang, Y, Han, L, Zheng, J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.555 Å) | | Cite: | Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins
Nat.Struct.Mol.Biol., 23, 2016
|
|
6IRQ
 
 | |
6JDG
 
 | | Complexed crystal structure of PaSSB with ssDNA dT20 at 2.39 angstrom resolution | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Single-stranded DNA-binding protein | | Authors: | Huang, Y.H, Huang, C.Y. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Complexed crystal structure of SSB reveals a novel single-stranded DNA binding mode (SSB)3:1: Phe60 is not crucial for defining binding paths.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
3MJ0
 
 | |
5YYU
 
 | |
2FMM
 
 | | Crystal Structure of EMSY-HP1 complex | | Descriptor: | Chromobox protein homolog 1, Protein EMSY, SULFATE ION | | Authors: | Huang, Y. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the HP1-EMSY complex reveals an unusual mode of HP1 binding.
Structure, 14, 2006
|
|
4JC2
 
 | | Isolation, Cloning and Biophysical Analysis of a Novel Hexameric Green Fluorescent Protein from a Philippine Soft Coral | | Descriptor: | asFP504 | | Authors: | Huang, Y.C, Emralino, F.L, Liu, F.C, Saloma, C.P, Bascos, N.A, Chen, C.J. | | Deposit date: | 2013-02-21 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Isolation, Cloning and Biophysical Analysis of a Novel Hexameric Green Fluorescent Protein from a Philippine Soft Coral
To be Published
|
|
1D2H
 
 | | CRYSTAL STRUCTURE OF R175K MUTANT GLYCINE N-METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLHOMOCYSTEINE | | Descriptor: | GLYCINE N-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Huang, Y, Komoto, J, Takusagawa, F, Konishi, K, Takata, Y. | | Deposit date: | 1999-10-11 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
5YUN
 
 | | Crystal structure of SSB complexed with myc | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Single-stranded DNA-binding protein | | Authors: | Huang, Y.H, Huang, C.Y. | | Deposit date: | 2017-11-22 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of SSB complexed with inhibitor myricetin.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6AEQ
 
 | |
2HKY
 
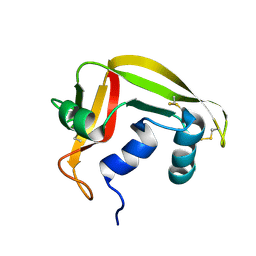 | | NMR solution structure of human RNase 7 | | Descriptor: | Ribonuclease 7 | | Authors: | Huang, Y.-C, Chen, C, Lou, Y.-C. | | Deposit date: | 2006-07-06 | | Release date: | 2006-12-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity.
J.Biol.Chem., 282, 2007
|
|
1D2G
 
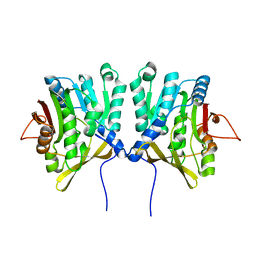 | | CRYSTAL STRUCTURE OF R175K MUTANT GLYCINE N-METHYLTRANSFERASE FROM RAT LIVER | | Descriptor: | GLYCINE N-METHYLTRANSFERASE | | Authors: | Huang, Y, Komoto, J, Takusagawa, F, Konishi, K, Takata, Y. | | Deposit date: | 1999-10-08 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
1D2C
 
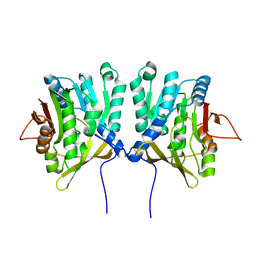 | | METHYLTRANSFERASE | | Descriptor: | PROTEIN (GLYCINE N-METHYLTRANSFERASE) | | Authors: | Huang, Y, Takusagawa, F. | | Deposit date: | 1999-09-23 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
4QXW
 
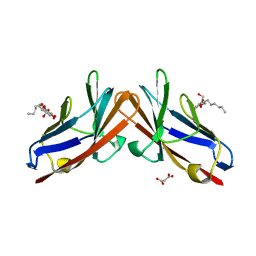 | | Crystal structure of the human CEACAM1 membrane distal amino terminal (N)-domain | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Huang, Y.H, Gandhi, A.K, Russell, A, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2014-07-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
8GTD
 
 | | Cryo-EM model of the marine siphophage vB_DshS-R4C portal-adaptor complex | | Descriptor: | Head-to-tail joining protein, Portal protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTB
 
 | | Cryo-EM structure of the marine siphophage vB_DshS-R4C tail tube protein | | Descriptor: | Major tail protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTC
 
 | | Cryo-EM model of the marine siphophage vB_DshS-R4C baseplate-tail complex | | Descriptor: | Distal tail protein, Hub protein, Major tail protein, ... | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTF
 
 | | Cryo-EM model of the marine siphophage vB_DshS-R4C stopper-terminator complex | | Descriptor: | Head-to-tail joining protein, Major tail protein, Terminator protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
2GFA
 
 | | double tudor domain complex structure | | Descriptor: | Jumonji domain-containing protein 2A, peptide | | Authors: | Huang, Y, Fang, J, Bedford, M.T, Zhang, Y, Xu, R.M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A
Science, 312, 2006
|
|
8WXS
 
 | |
2GF7
 
 | | Double tudor domain structure | | Descriptor: | Jumonji domain-containing protein 2A, SULFATE ION | | Authors: | Huang, Y, Fang, J, Bedford, M.T, Zhang, Y, Xu, R.M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A
Science, 312, 2006
|
|
9L2G
 
 | | Structure of SARM1 2C-mutant bound to M1 | | Descriptor: | NAD(+) hydrolase SARM1, [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(3-sulfanylpyridin-1-yl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Huang, Y, Zhang, J, Zheng, S, Wang, X. | | Deposit date: | 2024-12-17 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Stepwise Activation of SARM1 by a Biosynthetic NMN mimic
To Be Published
|
|
9L2D
 
 | | Structure of SARM1 bound to M1 in the intermediate state 1 | | Descriptor: | NAD(+) hydrolase SARM1, [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(3-sulfanylpyridin-1-yl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Huang, Y, Zhang, J, Zheng, S, Wang, X. | | Deposit date: | 2024-12-17 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Stepwise Activation of SARM1 by a Biosynthetic NMN mimic
To Be Published
|
|
