3TJC
 
 | | Co-crystal structure of jak2 with thienopyridine 8 | | Descriptor: | 4-amino-N-methyl-2-[4-(morpholin-4-yl)phenyl]thieno[3,2-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Huang, X. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent and highly selective thienopyridine janus kinase 2 inhibitors.
J.Med.Chem., 54, 2011
|
|
3TJD
 
 | | co-crystal structure of Jak2 with thienopyridine 19 | | Descriptor: | 4-amino-2-[4-(tert-butylsulfamoyl)phenyl]-N-methylthieno[3,2-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Huang, X. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of potent and highly selective thienopyridine janus kinase 2 inhibitors.
J.Med.Chem., 54, 2011
|
|
8IGO
 
 | | Crystal structure of apo SARS-CoV-2 main protease | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
3JZK
 
 | | crystal structure of MDM2 with chromenotriazolopyrimidine 1 | | Descriptor: | (6R,7S)-6,7-bis(4-bromophenyl)-7,11-dihydro-6H-chromeno[4,3-d][1,2,4]triazolo[1,5-a]pyrimidine, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and optimization of chromenotriazolopyrimidines as potent inhibitors of the mouse double minute 2-tumor protein 53 protein-protein interaction.
J.Med.Chem., 52, 2009
|
|
4HBM
 
 | | Ordering of the N Terminus of Human MDM2 by Small Molecule Inhibitors | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S)-1-hydroxybutan-2-yl]-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ordering of the N-terminus of human MDM2 by small molecule inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
4HNF
 
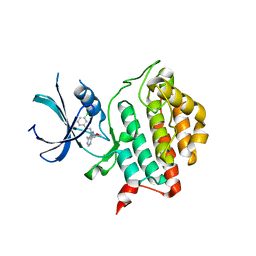 | | Crystal structure of ck1d in complex with pf4800567 | | Descriptor: | 3-[(3-chlorophenoxy)methyl]-1-(tetrahydro-2H-pyran-4-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Casein kinase I isoform delta | | Authors: | Huang, X, Long, A.M, Zhao, H. | | Deposit date: | 2012-10-19 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for the potent and selective inhibition of casein kinase 1 epsilon.
J.Med.Chem., 55, 2012
|
|
4HOK
 
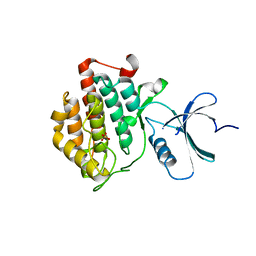 | | crystal structure of apo ck1e | | Descriptor: | Casein kinase I isoform epsilon, SULFATE ION | | Authors: | Huang, X, Long, A.M, Zhao, H. | | Deposit date: | 2012-10-22 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for the potent and selective inhibition of casein kinase 1 epsilon.
J.Med.Chem., 55, 2012
|
|
4HNI
 
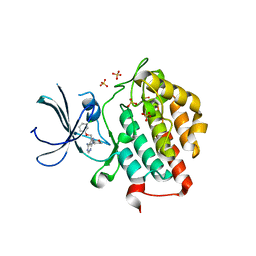 | | crystal structure of ck1e in complex with PF4800567 | | Descriptor: | 3-[(3-chlorophenoxy)methyl]-1-(tetrahydro-2H-pyran-4-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Casein kinase I isoform epsilon, SULFATE ION | | Authors: | Huang, X, Long, A.M, Zhao, H. | | Deposit date: | 2012-10-19 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis for the potent and selective inhibition of casein kinase 1 epsilon.
J.Med.Chem., 55, 2012
|
|
4I9I
 
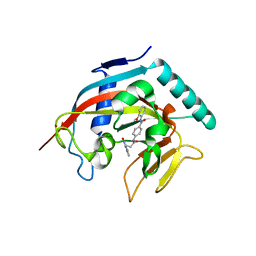 | | Crystal structure of tankyrase 1 with compound 4 | | Descriptor: | N-(2-methoxyphenyl)-4-{[3-(4-oxo-3,4-dihydroquinazolin-2-yl)propanoyl]amino}benzamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2012-12-05 | | Release date: | 2013-02-06 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a class of novel tankyrase inhibitors that bind to both the nicotinamide pocket and the induced pocket.
J.Med.Chem., 56, 2013
|
|
4RPV
 
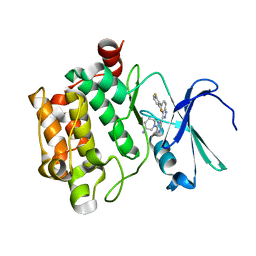 | | co-crystal structure of Pim1 with compound 3 | | Descriptor: | (3S)-1-{6-[5-(2,6-difluorophenyl)-2H-indazol-3-yl]pyrazin-2-yl}piperidin-3-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Huang, X. | | Deposit date: | 2014-10-31 | | Release date: | 2015-02-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The discovery of novel 3-(pyrazin-2-yl)-1H-indazoles as potent pan-Pim kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2OFU
 
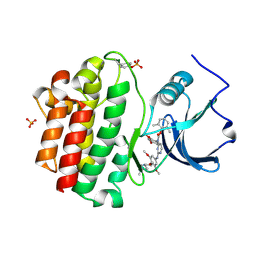 | | x-ray crystal structure of 2-aminopyrimidine carbamate 43 bound to Lck | | Descriptor: | 2,6-DIMETHYLPHENYL 2-(3,5-DIMETHOXY-4-(3-(4-METHYLPIPERAZIN-1-YL)PROPOXY)PHENYLAMINO)PYRIMIDIN- 4-YL(2,4-DIMETHOXYPHENYL)CARBAMATE, Proto-oncogene tyrosine-protein kinase LCK, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel 2-Aminopyrimidine Carbamates as Potent and Orally Active Inhibitors of Lck: Synthesis, SAR, and in Vivo Antiinflammatory Activity
J.Med.Chem., 49, 2006
|
|
2OFV
 
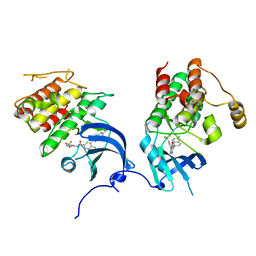 | | crystal structure of aminoquinazoline 1 bound to Lck | | Descriptor: | 3-(2-AMINOQUINAZOLIN-6-YL)-4-METHYL-N-[3-(TRIFLUOROMETHYL)PHENYL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-04 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
2OG8
 
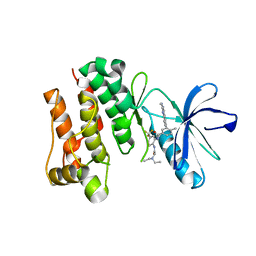 | | crystal structure of aminoquinazoline 36 bound to Lck | | Descriptor: | N-{2-[(N,N-DIETHYLGLYCYL)AMINO]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[2-(METHYLAMINO)QUINAZOLIN-6-YL]BENZAMIDE, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Aminoquinazolines as Potent, Orally Bioavailable Inhibitors of Lck: Synthesis, SAR, and in Vivo Anti-Inflammatory Activity
J.Med.Chem., 49, 2006
|
|
6LTU
 
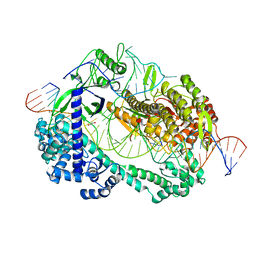 | | Crystal structure of Cas12i2 ternary complex with double Mg2+ bound in catalytic pocket | | Descriptor: | 1,2-ETHANEDIOL, Cas12i2, DNA (35-MER), ... | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
6LTR
 
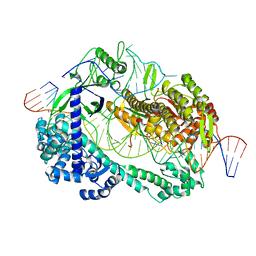 | | Crystal structure of Cas12i2 ternary complex with single Mg2+ bound in catalytic pocket | | Descriptor: | 1,2-ETHANEDIOL, Cas12i2, DNA (35-MER), ... | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
6LU0
 
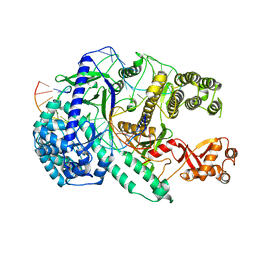 | | Crystal structure of Cas12i2 ternary complex with 12 nt spacer | | Descriptor: | Cas12i2, DNA (5'-D(*GP*CP*CP*GP*CP*TP*TP*TP*CP*TP*T)-3'), DNA (5'-D(*GP*CP*TP*TP*GP*CP*TP*CP*TP*GP*TP*TP*GP*AP*AP*AP*GP*CP*GP*GP*C)-3'), ... | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-24 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
6LTP
 
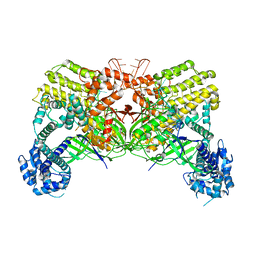 | | Crystal structure of Cas12i2 binary complex | | Descriptor: | Cas12i2, crRNA (56-mer RNA) | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
2JXW
 
 | |
6PZ4
 
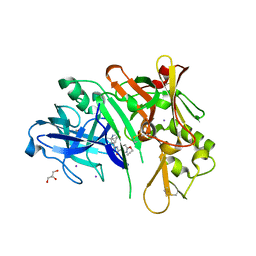 | | co-crystal structure of BACE with inhibitor AM-6494 | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Huang, X. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-23 | | Last modified: | 2020-03-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of AM-6494: A Potent and Orally Efficacious beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitor with in Vivo Selectivity over BACE2.
J.Med.Chem., 63, 2020
|
|
8I3A
 
 | |
8I3B
 
 | |
8I39
 
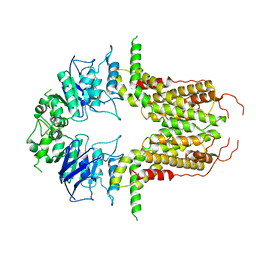 | | Cryo-EM structure of abscisic acid transporter AtABCG25 with ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Huang, X, Zhang, X, Zhang, P. | | Deposit date: | 2023-01-16 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure and molecular mechanism of abscisic acid transporter ABCG25.
Nat.Plants, 9, 2023
|
|
8I3C
 
 | |
8I38
 
 | |
