6LBA
 
 | | Cryo-EM structure of the AtMLKL2 tetramer | | Descriptor: | Protein kinase family protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
3M61
 
 | | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1 W3A | | Authors: | Jiang, L, Yuan, C, Wind, T, Andreasen, P.A, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition
TO BE PUBLISHED
|
|
6L04
 
 | | Crystal structure of uPA_H99Y in complex with 31F | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(2,4-dimethoxypyrimidin-5-yl)pyrazine-2-carboxamide, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of uPA_H99Y in complex with 31F
To Be Published
|
|
8BG9
 
 | | Murine amyloid-beta filaments with the Arctic mutation (E22G) from APP(NL-G-F) mouse brains | ABeta | | Descriptor: | Amyloid-beta protein 40 | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordber, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
8BG0
 
 | | Amyloid-beta tetrameric filaments with the Arctic mutation (E22G) from Alzheimer's disease brains | ABeta40 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordber, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
8BFZ
 
 | | Amyloid-beta 42 filaments extracted from the human brain with Arctic mutation (E22G) of Alzheimer's disease | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordberg, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
1Y8X
 
 | | Structural basis for recruitment of Ubc12 by an E2-binding domain in NEDD8's E1 | | Descriptor: | Ubiquitin-activating enzyme E1C, Ubiquitin-conjugating enzyme E2 M | | Authors: | Huang, D.T, Paydar, A, Zhuang, M, Waddell, M.B, Holton, J.M, Schulman, B.A. | | Deposit date: | 2004-12-13 | | Release date: | 2005-02-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1.
Mol.Cell, 17, 2005
|
|
5LHP
 
 | | The p-aminobenzamidine active site inhibited catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment, P-AMINO BENZAMIDINE, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P.J, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
5LHN
 
 | | The catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment Nb7, SULFATE ION, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
5LHQ
 
 | | The EGR-cmk active site inhibited catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment Nb7, L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P.J, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
5LHS
 
 | | The ligand free catalytic domain of murine urokinase-type plasminogen activator | | Descriptor: | NICKEL (II) ION, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P.J, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.047 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
5LHR
 
 | | The catalytic domain of murine urokinase-type plasminogen activator in complex with the active site binding inhibitory nanobody Nb22 | | Descriptor: | Camelid-Derived Antibody Fragment Nb22, Urokinase-type plasminogen activator | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P.J, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
2NVU
 
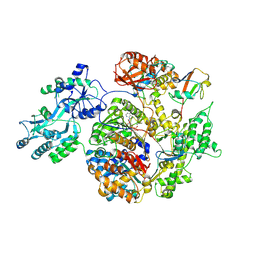 | | Structure of APPBP1-UBA3~NEDD8-NEDD8-MgATP-Ubc12(C111A), a trapped ubiquitin-like protein activation complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Maltose binding protein/NEDD8-activating enzyme E1 catalytic subunit chimera, ... | | Authors: | Huang, D.T, Hunt, H.W, Zhuang, M, Ohi, M.D, Holton, J.M, Schulman, B.A. | | Deposit date: | 2006-11-13 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity.
Nature, 445, 2007
|
|
2FA9
 
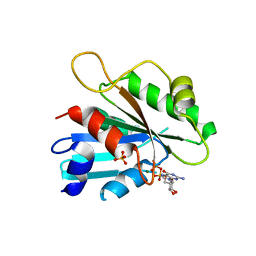 | | The crystal structure of Sar1[H79G]-GDP provides insight into the coat-controlled GTP hydrolysis in the disassembly of COP II | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rao, Y, Huang, M, Yuan, C, Bian, C, Hou, X. | | Deposit date: | 2005-12-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Sar1[H79G]-GDP Which Provides Insight into the Coat-controlled GTP Hydrolysis in the Disassembly of COP II
Chin.J.Struct.Chem., 25, 2006
|
|
2FMX
 
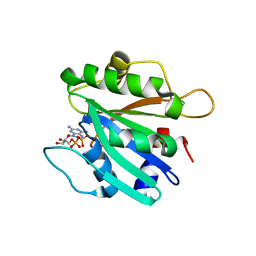 | | An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg(2+) | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rao, Y, Bian, C, Yuan, C, Li, Y, Huang, M. | | Deposit date: | 2006-01-10 | | Release date: | 2006-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg(2+)
Biochem.Biophys.Res.Commun., 348, 2006
|
|
1GND
 
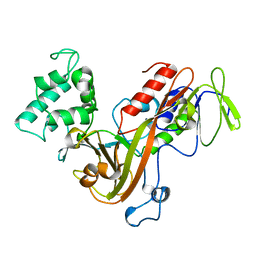 | | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, ALPHA-ISOFORM | | Descriptor: | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR | | Authors: | Schalk, I, Zeng, K, Wu, S.-K, Stura, E.A, Metteson, J, Huang, M, Tandon, A, Wilson, I.A, Balch, W.E. | | Deposit date: | 1996-07-10 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and mutational analysis of Rab GDP-dissociation inhibitor.
Nature, 381, 1996
|
|
3B9L
 
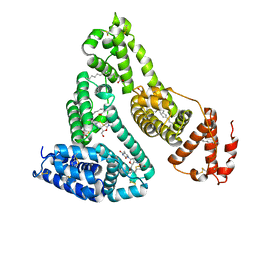 | | Human serum albumin complexed with myristate and AZT | | Descriptor: | 3'-azido-3'-deoxythymidine, MYRISTIC ACID, Serum albumin | | Authors: | Zhu, L, Yang, F, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography
J.Struct.Biol., 162, 2008
|
|
3B9M
 
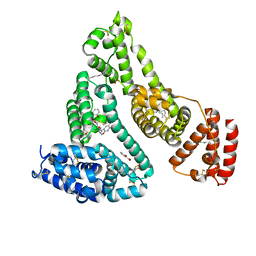 | | Human serum albumin complexed with myristate, 3'-azido-3'-deoxythymidine (AZT) and salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, 3'-azido-3'-deoxythymidine, MYRISTIC ACID, ... | | Authors: | Zhu, L, Yang, F, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography
J.Struct.Biol., 162, 2008
|
|
3G7N
 
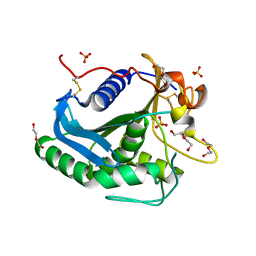 | | Crystal Structure of a Triacylglycerol Lipase from Penicillium Expansum at 1.3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lipase, PENTAETHYLENE GLYCOL, ... | | Authors: | Bian, C.B, Yuan, C, Chen, L.Q, Edward, J.M, Lin, L, Jiang, L.G, Huang, Z.X, Huang, M.D. | | Deposit date: | 2009-02-10 | | Release date: | 2010-02-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a triacylglycerol lipase from Penicillium expansum at 1.3 A determined by sulfur SAD
Proteins, 78, 2010
|
|
8Y6F
 
 | |
3MWI
 
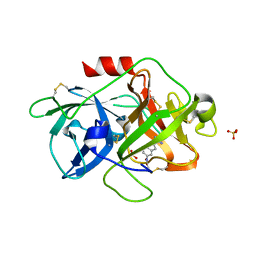 | | The complex crystal Structure of Urokianse and 5-nitro-1H-indole-2-amidine | | Descriptor: | 5-nitro-1H-indole-2-carboximidamide, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Yu, H.Y, Yuan, C, Huang, Z.X, Huang, M.D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The complex crystal Structure of Urokianse and 5-nitro-1H-indole-2-amidine
To be Published
|
|
4MD6
 
 | | Crystal structure of PDE5 in complex with inhibitor 5R | | Descriptor: | 3-(4-hydroxybenzyl)-1-(thiophen-2-yl)chromeno[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Cui, W, Huang, M, Shao, Y, Luo, H. | | Deposit date: | 2013-08-22 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 3-(4-hydroxybenzyl)-1-(thiophen-2-yl)chromeno[2,3-c]pyrrol-9(2H)-one as a phosphodiesterase-5 inhibitor and its complex crystal structure.
Biochem Pharmacol, 89, 2014
|
|
1XNZ
 
 | | Crystal Structure of Mn(II) form of E. coli. Methionine Aminopeptidase in complex with 5-(2-chlorophenyl)furan-2-carboxylic acid | | Descriptor: | 5-(2-CHLOROPHENYL)FURAN-2-CARBOXYLIC ACID, MANGANESE (II) ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.-Z, Xie, S.-X, Huang, M, Huang, W.-J, Lu, J.-P, Ma, Z.-Q. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Metalloform-Selective Inhibitors of Escherichia coli Methionine Aminopeptidase and X-ray Structure of a Mn(II)-Form Enzyme Complexed with an Inhibitor.
J.Am.Chem.Soc., 126, 2004
|
|
2FAT
 
 | | An anti-urokinase plasminogen activator receptor (UPAR) antibody: Crystal structure and binding epitope | | Descriptor: | FAB ATN-615, heavy chain, light chain | | Authors: | Li, Y, Parry, G, Shi, X, Chen, L, Callahan, J.A, Mazar, A.P, Huang, M. | | Deposit date: | 2005-12-07 | | Release date: | 2006-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | An anti-urokinase plasminogen activator receptor (uPAR) antibody: crystal structure and binding epitope
J.Mol.Biol., 365, 2007
|
|
3MHW
 
 | | The complex crystal Structure of Urokianse and 2-Aminobenzothiazole | | Descriptor: | 1,3-benzothiazol-2-amine, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L.-G, Yuan, C, Chen, L.-Q, Huang, M.-D. | | Deposit date: | 2010-04-09 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
