5HYC
 
 | | Structure based function annotation of a hypothetical protein MGG_01005 related to the development of rice blast fungus | | Descriptor: | Cytoplasmic dynein 1 intermediate chain 2, Uncharacterized protein | | Authors: | Liu, J, Li, G, Huang, J, Peng, Y.-l. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure based function-annotation of hypothetical protein MGG_01005 from Magnaporthe oryzae reveals it is the dynein light chain orthologue of dynlt1/3.
Sci Rep, 8, 2018
|
|
4G6F
 
 | | Crystal Structure of 10E8 Fab in Complex with an HIV-1 gp41 Peptide | | Descriptor: | 10E8 Heavy Chain, 10E8 Light Chain, gp41 MPER Peptide | | Authors: | Ofek, G, Huang, J, Connors, M, Kwong, P.D. | | Deposit date: | 2012-07-19 | | Release date: | 2012-09-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Broad and potent neutralization of HIV-1 by a gp41-specific human antibody.
Nature, 491, 2012
|
|
6LZL
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with Piperine | | Descriptor: | (2E,4E)-5-(2H-1,3-benzodioxol-5-yl)-1-(piperidin-1-yl)penta-2,4-dien-1-one, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Liu, Z.H, Wu, D, Lu, W.Q, Huang, J. | | Deposit date: | 2020-02-19 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with Piperine
To Be Published
|
|
2XCS
 
 | | The 2.1A crystal structure of S. aureus Gyrase complex with GSK299423 and DNA | | Descriptor: | 5'-5UA*D(GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP *CP*TP*AP*CP*GP*GP*CP*T)-3', 6-METHOXY-4-(2-{4-[([1,3]OXATHIOLO[5,4-C]PYRIDIN-6-YLMETHYL)AMINO]PIPERIDIN-1-YL}ETHYL)QUINOLINE-3-CARBONITRILE, DNA GYRASE SUBUNIT B, ... | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCT
 
 | | The twinned 3.35A structure of S. aureus Gyrase complex with Ciprofloxacin and DNA | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, 5'-D(AP*GP*CP*CP*GP*TP*AP*G)-3', 5'-D(GP*TP*AP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3', ... | | Authors: | Bax, B.D, Chan, P, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCR
 
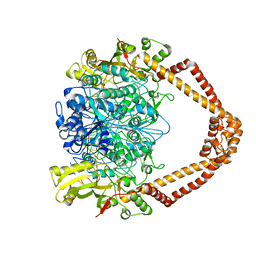 | | The 3.5A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase complexed with GSK299423 and DNA | | Descriptor: | 5'-D(*5UA*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP *GP*CP*TP)-3', 5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP *GP*CP*TP)-3', 6-METHOXY-4-(2-{4-[([1,3]OXATHIOLO[5,4-C]PYRIDIN-6-YLMETHYL)AMINO]PIPERIDIN-1-YL}ETHYL)QUINOLINE-3-CARBONITRILE, ... | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCO
 
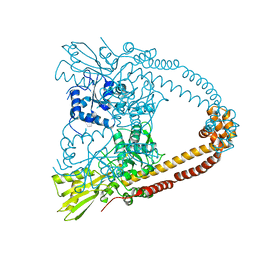 | | The 3.1A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | CALCIUM ION, DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XCQ
 
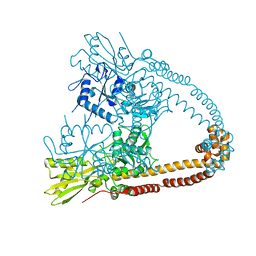 | | The 2.98A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
2XKJ
 
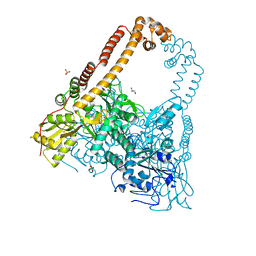 | | CRYSTAL STRUCTURE OF CATALYTIC CORE OF A. BAUMANNII TOPO IV (PARE- PARC FUSION TRUNCATE) | | Descriptor: | GLYCEROL, SULFATE ION, TOPOISOMERASE IV | | Authors: | Wohlkonig, A, Chan, P.F, Fosberry, A.P, Homes, P, Huang, J, Kranz, M, Leydon, V.R, Miles, T.J, Pearson, N.D, Perera, R.L, Shillings, A.J, Gwynn, M.N, Bax, B.D. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Quinolone Inhibition of Type Iia Topoisomerases and Target-Mediated Resistance
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XKK
 
 | | CRYSTAL STRUCTURE OF MOXIFLOXACIN, DNA, and A. BAUMANNII TOPO IV (PARE-PARC FUSION TRUNCATE) | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA, MAGNESIUM ION, ... | | Authors: | Wohlkonig, A, Chan, P.F, Fosberry, A.P, Homes, P, Huang, J, Kranz, M, Leydon, V.R, Miles, T.J, Pearson, N.D, Perera, R.L, Shillings, A.J, Gwynn, M.N, Bax, B.D. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis of Quinolone Inhibition of Type Iia Topoisomerases and Target-Mediated Resistance
Nat.Struct.Mol.Biol., 17, 2010
|
|
8W68
 
 | | Crystal structure of Q9PR55 at pH 6.0 (use NMR model) | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-08-28 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWC
 
 | | Crystal structure of Q9PR55 at pH 6.0 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWA
 
 | | Crystal structure of Q9PR55 at pH 6.5 | | Descriptor: | SULFATE ION, Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWB
 
 | | Crystal structure of Q9PR55 at pH 7.5 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8WKS
 
 | | Cryo-EM structure of DSR2-TUBE complex | | Descriptor: | SIR2-like domain-containing protein, TUBE | | Authors: | Gao, A, Huang, J, Zhu, K. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
9AYJ
 
 | | Cryo-EM structure of human Cav3.2 with TTA-P2 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,5-dichloro-N-[(1-{[(4S)-2,2-dimethyloxan-4-yl]methyl}-4-fluoropiperidin-4-yl)methyl]benzamide, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYK
 
 | | Cryo-EM structure of human Cav3.2 with ML218 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,5-dichloro-N-{[(1R,5S,6r)-3-(3,3-dimethylbutyl)-3-azabicyclo[3.1.0]hexan-6-yl]methyl}benzamide, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
8WKX
 
 | | Cryo-EM structure of DSR2 | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Gao, A, Huang, J, Zhu, K. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
8WKT
 
 | | Cryo-EM structure of DSR2-DSAD1 complex | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Gao, A, Huang, J, Zhu, K. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
9AYL
 
 | | Cryo-EM structure of human Cav3.2 with ACT-709478 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYG
 
 | | Cryo-EM structure of apo state human Cav3.2 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYH
 
 | | Cryo-EM structure of human Cav3.2 with TTA-A2 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(4-cyclopropylphenyl)-N-{(1R)-1-[5-(2,2,2-trifluoroethoxy)pyridin-2-yl]ethyl}acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
8S9B
 
 | | Cryo-EM structure of Nav1.7 with LCM | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8S9C
 
 | | Cryo-EM structure of Nav1.7 with CBZ | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
4GP3
 
 | | The crystal structure of human fascin 1 K358A mutant | | Descriptor: | BROMIDE ION, CHLORIDE ION, Fascin, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
