6KIJ
 
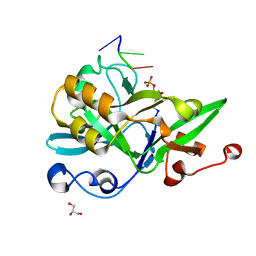 | | Crystal structure of yedK with ssDNA containing an abasic site | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*CP*GP*TP*CP*G)-3'), GLYCEROL, PENTANE-3,4-DIOL-5-PHOSPHATE, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-07-18 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Molecular basis of abasic site sensing in single-stranded DNA by the SRAP domain of E. coli yedK.
Nucleic Acids Res., 47, 2019
|
|
6J4U
 
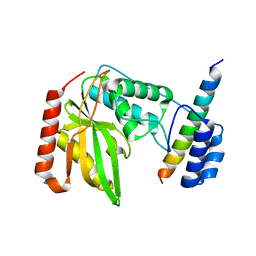 | |
6J4Q
 
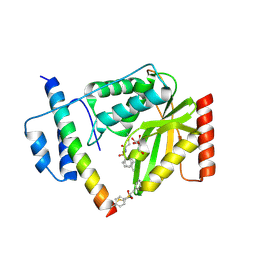 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | Descriptor: | GLYCEROL, N-[(2S)-4-chloro-3-oxo-1-phenyl-butan-2-yl]-4-methyl-benzenesulfonamide, Small vasohibin-binding protein, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6KBZ
 
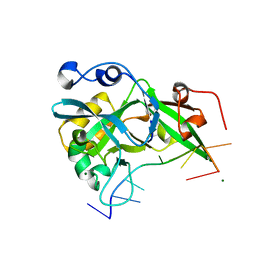 | |
6KBX
 
 | |
7V1M
 
 | |
7V1L
 
 | |
7V1K
 
 | | Apo structure of sNASP core | | Descriptor: | Isoform 2 of Nuclear autoantigenic sperm protein | | Authors: | Bao, H, Huang, H. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.287 Å) | | Cite: | NASP maintains histone H3-H4 homeostasis through two distinct H3 binding modes.
Nucleic Acids Res., 50, 2022
|
|
2OMJ
 
 | | solution structure of LARG PDZ domain | | Descriptor: | Rho guanine nucleotide exchange factor 12 | | Authors: | Liu, J, Huang, H, Hu, Q. | | Deposit date: | 2007-01-22 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | solution structure and dynamics of the LARG PDZ domain
To be Published
|
|
2OS6
 
 | |
2AX5
 
 | | Solution Structure of Urm1 from Saccharomyces Cerevisiae | | Descriptor: | Hypothetical 11.0 kDa protein in FAA3-MAS3 intergenic region | | Authors: | Xu, J, Huang, H, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2005-09-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 and its implications for the origin of protein modifiers.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7CJM
 
 | | SARS CoV-2 PLpro in complex with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Fu, Z, Huang, H. | | Deposit date: | 2020-07-11 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery.
Nat Commun, 12, 2021
|
|
2KXJ
 
 | | Solution structure of UBX domain of human UBXD2 protein | | Descriptor: | UBX domain-containing protein 4 | | Authors: | Wu, Q, Huang, H, Zhang, J, Hu, Q, Wu, J, Shi, Y. | | Deposit date: | 2010-05-06 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution strcture of UBX domain of human UBXD2 protein
To be Published
|
|
7CJ0
 
 | |
7CIZ
 
 | |
7DSY
 
 | | Crystal Structure of RNase L in complex with KM05073 | | Descriptor: | 1-chloranyl-3-methylsulfinyl-6,7-dihydro-5H-2-benzothiophen-4-one, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, PHOSPHATE ION, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2021-01-03 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
7DTS
 
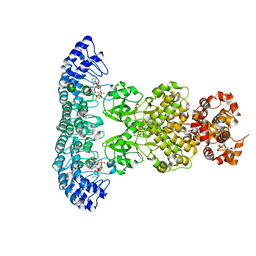 | | Crystal structure of RNase L in complex with AC40357 | | Descriptor: | 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2021-01-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
7ELW
 
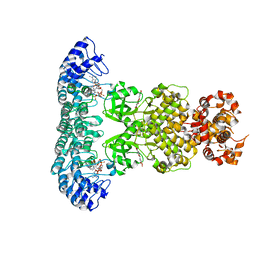 | | Crystal structure of RNase L in complex with Myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2021-04-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
5TVT
 
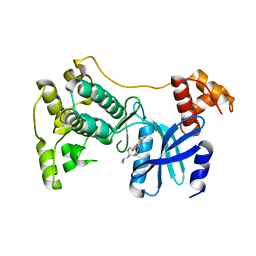 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 9-(3,5-dichloro-4-hydroxyphenyl)-1-{trans-4-[(dimethylamino)methyl]cyclohexyl}-3-methyl-3,4-dihydropyrimido[5,4-c][1,5]naphthyridin-2(1H)-one, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-Y, Huang, H, Gray, N.S, Dhe-Paganon, S. | | Deposit date: | 2016-11-10 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of Maternal Embryonic Leucine Zipper Kinase
To Be Published
|
|
7DG6
 
 | |
6O04
 
 | | M.tb MenD IntII bound with Inhibitor | | Descriptor: | (1~{R},2~{S},5~{S},6~{S})-2-[(1~{S})-1-[3-[(4-azanylidene-2-methyl-1~{H}-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl (phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-3-ium-2-yl]-1,4-bis(oxidanyl)-4-oxidanylidene-butyl]-6-oxidanyl-5-(3-oxid anyl-3-oxidanylidene-prop-1-en-2-yl)oxy-cyclohex-3-ene-1-carboxylic acid, 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Johnston, J.M, Bashiri, G, Bulloch, E.M, Jirgis, E.M.N, Nigon, L.V, Chuang, H, Baker, E.N. | | Deposit date: | 2019-02-15 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
6O0G
 
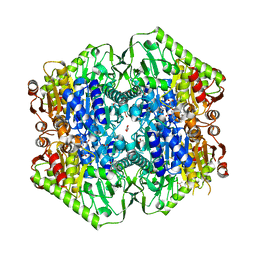 | | M.tb MenD bound to Intermediate I and Inhibitor | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, 2-OXOGLUTARIC ACID, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Johnston, J.M, Bashiri, G, Bulloch, E.M.M, Jirgis, E.M.N, Chuang, H, Nigon, L.V, Baker, E.N. | | Deposit date: | 2019-02-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
7FA3
 
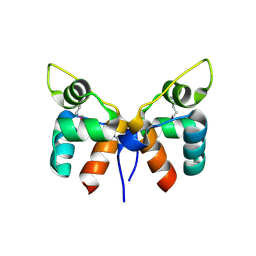 | |
6O0J
 
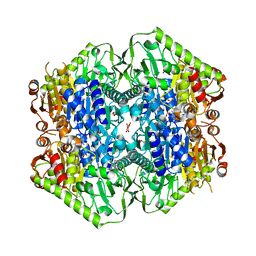 | | M.tb MenD with ThDP and Inhibitor bound | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ACETATE ION, ... | | Authors: | Johnston, J.M, Bashiri, G, Bulloch, E.M.M, Jirgis, E.M.N, Nigon, L.V, Chuang, H, Ho, N.A.T, Baker, E.N. | | Deposit date: | 2019-02-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
2L26
 
 | | Rv0899 from Mycobacterium tuberculosis contains two separated domains | | Descriptor: | Uncharacterized protein Rv0899/MT0922 | | Authors: | Shi, C, Li, J, Gao, Y, Wu, K, Huang, H, Tian, C. | | Deposit date: | 2010-08-12 | | Release date: | 2011-08-17 | | Last modified: | 2011-12-07 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of Mycobacterium tuberculosis Rv0899 Reveal a Monomeric Membrane-Anchoring Protein with Two Separate Domains
J.Mol.Biol., 2011
|
|
