8XBF
 
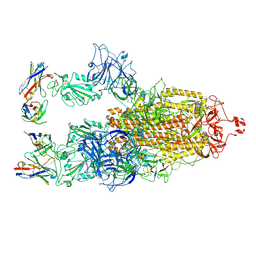 | | Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, O5C2, heavy chain, ... | | Authors: | Hsu, H.F, Wu, M.H, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody.
JCI Insight, 9, 2024
|
|
8G6E
 
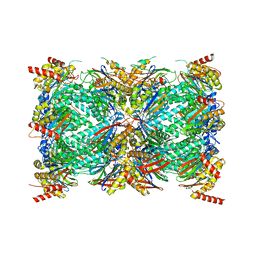 | | Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304 | | Descriptor: | (7S,10S,13S)-N-cyclopentyl-10-[2-(morpholin-4-yl)ethyl]-9,12-dioxo-13-(2-oxopyrrolidin-1-yl)-2-oxa-8,11-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-7-carboxamide, Proteasome subunit alpha type, Proteasome subunit alpha type-1, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
8FZ6
 
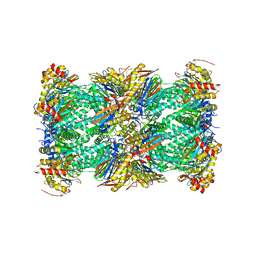 | | The human PI31 complexed with bovine 20S proteasome | | Descriptor: | Proteasome inhibitor PI31 subunit, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Eta igh-resolution structure of mammalian PI31-20S proteasome complex reveals mechanism of proteasome inhibition.
J.Biol.Chem., 299, 2023
|
|
8FZ5
 
 | | The PI31-free Bovine 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Eta igh-resolution structure of mammalian PI31-20S proteasome complex reveals mechanism of proteasome inhibition.
J.Biol.Chem., 299, 2023
|
|
6WNK
 
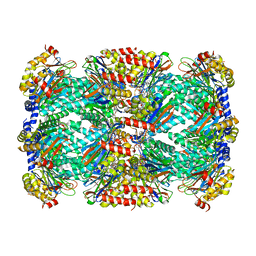 | | Macrocyclic peptides TDI5575 that selectively inhibit the Mycobacterium tuberculosis proteasome | | Descriptor: | (12S,15S)-N-[(2-fluorophenyl)methyl]-10,13-dioxo-12-{2-oxo-2-[(2R)-2-phenylpyrrolidin-1-yl]ethyl}-2-oxa-11,14-diazatricyclo[15.2.2.1~3,7~]docosa-1(19),3(22),4,6,17,20-hexaene-15-carboxamide, CITRIC ACID, DIMETHYLFORMAMIDE, ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Macrocyclic Peptides that Selectively Inhibit the Mycobacterium tuberculosis Proteasome.
J.Med.Chem., 64, 2021
|
|
7RPQ
 
 | |
7RQF
 
 | |
7RQH
 
 | |
8SXH
 
 | |
8SXG
 
 | |
8SXE
 
 | |
8SXF
 
 | |
8UD9
 
 | | Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304 | | Descriptor: | (7S,10S,13S)-N-cyclopentyl-10-[2-(morpholin-4-yl)ethyl]-9,12-dioxo-13-(2-oxopyrrolidin-1-yl)-2-oxa-8,11-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-7-carboxamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-09-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
5THO
 
 | | Crystal Structure of Mycobacterium Tuberculosis Proteasome in complex with N,C-capped Dipeptide Inhibitor PKS2205 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2016-09-30 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
6U3Q
 
 | | The atomic structure of a human adeno-associated virus capsid isolate (AAVhu69/AAVv66) | | Descriptor: | Capsid protein VP1 | | Authors: | Hsu, H.-L, Brown, A, Loveland, A, Tai, P, Korostelev, A, Gao, G. | | Deposit date: | 2019-08-22 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural characterization of a novel human adeno-associated virus capsid with neurotropic properties.
Nat Commun, 11, 2020
|
|
6C31
 
 | | Crystal structure of TetR family protein Rv0078 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*TP*TP*AP*CP*CP*GP*GP*CP*AP*GP*TP*CP*TP*GP*CP*TP*TP*GP*TP*AP*AP*A)-3'), DNA (5'-D(P*AP*CP*AP*AP*GP*CP*AP*GP*AP*CP*TP*GP*CP*CP*GP*GP*TP*AP*AP*C)-3'), TetR family transcriptional regulator | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2018-01-09 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cytokinin Signaling in Mycobacterium tuberculosis.
MBio, 9, 2018
|
|
8G6F
 
 | | Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs | | Descriptor: | (2S)-N-[(E,2S)-1-(1H-indol-3-yl)-4-methylsulfonyl-but-3-en-2-yl]-2-[[(2S)-3-(1H-indol-3-yl)-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]-4-methyl-pentanamide, Proteasome endopeptidase complex, Proteasome subunit alpha type-1, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
5TRS
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2144 | | Descriptor: | N-tert-butoxy-N~2~-(5-methyl-1,2-oxazole-3-carbonyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.083567 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5URA
 
 | | Enantiomer-Specific Binding of the Potent Antinociceptive Agent SBFI-26 to Anandamide transporters FABP7 | | Descriptor: | (1S,2S,3S,4S)-3-{[(naphthalen-1-yl)oxy]carbonyl}-2,4-diphenylcyclobutane-1-carboxylic acid, Fatty acid-binding protein, brain, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85002172 Å) | | Cite: | The Antinociceptive Agent SBFI-26 Binds to Anandamide Transporters FABP5 and FABP7 at Two Different Sites.
Biochemistry, 56, 2017
|
|
5TRR
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2169 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-N-[(naphthalen-1-yl)methyl]-L-alaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRY
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2206 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-3-methoxy-1-(naphthalen-1-ylmethylamino)-1-oxidanylidene-propan-2-yl]-4-oxidanylidene-2-(3-phenylpropanoylamino)-4-piperidin-1-yl-butanamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.000008 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TS0
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2208 | | Descriptor: | (2S)-N-{(2S)-3-methoxy-1-[(naphthalen-1-ylmethyl)amino]-1-oxopropan-2-yl}-4-oxo-2-[(3-phenylpropanoyl)amino]-4-(1H-pyrrol-1-yl)butanamide (non-preferred name), Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84679747 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5UR9
 
 | | Enantiomer-Specific Binding of the Potent Antinociceptive Agent SBFI-26 to Anandamide transporters FABP5 | | Descriptor: | (1S,2S,3S,4S)-3-{[(naphthalen-1-yl)oxy]carbonyl}-2,4-diphenylcyclobutane-1-carboxylic acid, Fatty acid-binding protein, epidermal, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19800353 Å) | | Cite: | The Antinociceptive Agent SBFI-26 Binds to Anandamide Transporters FABP5 and FABP7 at Two Different Sites.
Biochemistry, 56, 2017
|
|
5TRG
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide DPLG-2 | | Descriptor: | N,N-diethyl-N~2~-[(2E)-3-phenylprop-2-enoyl]-L-asparaginyl-4-fluoro-N-[(naphthalen-1-yl)methyl]-L-phenylalaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, R.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5WM9
 
 | |
