4X3U
 
 | | Crystal structure of chromobox homolog 7 (CBX7) chromodomain with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Chromobox protein homolog 7 | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
7Z9L
 
 | | Phen-DC3 intercalation causes hybrid-to-antiparallel transformation of human telomeric DNA G-quadruplex | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N2,N9-bis(1-methylquinolin-3-yl)-1,10-phenanthroline-2,9-dicarboxamide | | Authors: | Ghosh, A, Trajkovski, M, Teulade-Fichou, M.P, Gabelica, V, Plavec, J. | | Deposit date: | 2022-03-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Phen-DC 3 Induces Refolding of Human Telomeric DNA into a Chair-Type Antiparallel G-Quadruplex through Ligand Intercalation.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8UY6
 
 | | Aquaporin Z with ALFA tag and bound to nanobody | | Descriptor: | Aquaporin Z, CARDIOLIPIN, anti-ALFA nanobody | | Authors: | Stover, L, Bahramimoghaddam, H, Wang, L, Zhou, M, Laganowsky, A. | | Deposit date: | 2023-11-13 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Grafting the ALFA tag for structural studies of aquaporin Z.
J Struct Biol X, 9, 2024
|
|
4X3S
 
 | | Crystal structure of chromobox homology 7 (CBX7) with SETDB1-1170me3 Peptide | | Descriptor: | CITRIC ACID, Chromobox protein homolog 7, FE (III) ION, ... | | Authors: | Ren, C, Plotnikov, A.N, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
4X3T
 
 | | Crystal structure of chromobox homolog 7 (CBX7) chromodomain with MS37452 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(2,3-dimethoxybenzoyl)piperazin-1-yl]-2-(3-methylphenoxy)ethanone, Chromobox protein homolog 7, ... | | Authors: | Ren, C, Jakoncic, J, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
5ULA
 
 | |
7LHV
 
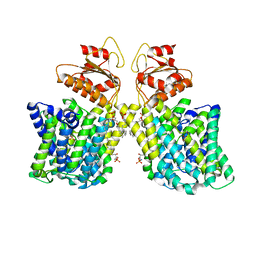 | | Structure of Arabidopsis thaliana sulfate transporter AtSULTR4;1 | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SULFATE ION, ... | | Authors: | Wang, L, Chen, K, Zhou, M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure and function of an Arabidopsis thaliana sulfate transporter.
Nat Commun, 12, 2021
|
|
8ERP
 
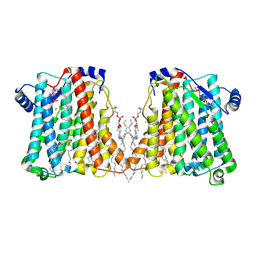 | | Structure of Xenopus cholinephosphotransferase1 in complex with CDP-choline | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Cholinephosphotransferase 1, MAGNESIUM ION, ... | | Authors: | Wang, L, Zhou, M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a eukaryotic cholinephosphotransferase-1 reveals mechanisms of substrate recognition and catalysis.
Nat Commun, 14, 2023
|
|
8ERO
 
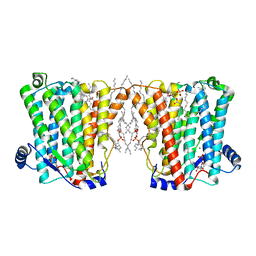 | | Structure of Xenopus cholinephosphotransferase1 in complex with CDP | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CYTIDINE-5'-DIPHOSPHATE, Cholinephosphotransferase 1, ... | | Authors: | Wang, L, Zhou, M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a eukaryotic cholinephosphotransferase-1 reveals mechanisms of substrate recognition and catalysis.
Nat Commun, 14, 2023
|
|
7S8U
 
 | |
5JPD
 
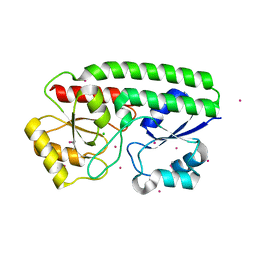 | | Metal ABC transporter from Listeria monocytogenes with cadmium | | Descriptor: | CADMIUM ION, CHLORIDE ION, Manganese-binding lipoprotein MntA | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-03 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Metal ABC transporter from Listeria monocytogenes with cadmium
to be published
|
|
5IWS
 
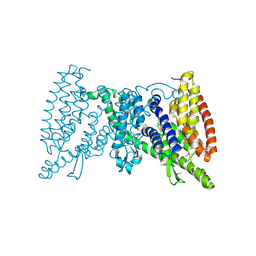 | | Crystal structure of the transporter MalT, the EIIC domain from the maltose-specific phosphotransferase system | | Descriptor: | Protein-N(Pi)-phosphohistidine-sugar phosphotransferase (Enzyme II of the phosphotransferase system) (PTS system glucose-specific IIBC component), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | McCoy, J.G, Ren, Z, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | The Structure of a Sugar Transporter of the Glucose EIIC Superfamily Provides Insight into the Elevator Mechanism of Membrane Transport.
Structure, 24, 2016
|
|
3DWH
 
 | |
8DL6
 
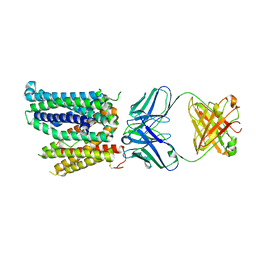 | | Cryo-EM structure of human ferroportin/slc40 bound to Ca2+ in nanodisc | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, CALCIUM ION, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of Ca 2+ transport by ferroportin.
Elife, 12, 2023
|
|
5JQC
 
 | | Crystal structure putative autolysin from Listeria monocytogenes | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Lmo1076 protein, ... | | Authors: | Chang, C, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-18 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal structure putative autolysin from Listeria monocytogenes
To Be Published
|
|
3EB4
 
 | | Voltage-dependent K+ channel beta subunit (I211R) in complex with cortisone | | Descriptor: | 17,21-DIHYDROXYPREGNA-1,4-DIENE-3,11,20-TRIONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Voltage-gated potassium channel subunit beta-2 | | Authors: | Pan, Y, Weng, J, Kabaleeswaran, V, Li, H, Cao, Y, Bhosle, R.C, Zhou, M. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cortisone dissociates the Shaker family K+ channels from their beta subunits.
Nat.Chem.Biol., 4, 2008
|
|
4JAL
 
 | | Crystal structure of tRNA (Um34/Cm34) methyltransferase TrmL from Escherichia coli with SAH | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Liu, R.J, Zhou, M, Wang, E.D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tRNA recognition mechanism of the minimalist SPOUT methyltransferase, TrmL
Nucleic Acids Res., 41, 2013
|
|
5U5S
 
 | |
6GBY
 
 | | Copper nitrite reductase from Achromobacter cycloclastes: non-polymorph separated dataset 1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2018-04-16 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6GBB
 
 | | Copper nitrite reductase from Achromobacter cycloclastes: large cell polymorph dataset 1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6GCG
 
 | | Copper nitrite reductase from Achromobacter cycloclastes: large polymorph dataset 15 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2018-04-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.80015242 Å) | | Cite: | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6GZW
 
 | |
6FNE
 
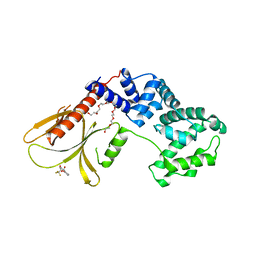 | | Structure of human Brag2 (Sec7-PH domains) with the inhibitor Bragsin bound to the PH domain | | Descriptor: | (2~{S})-6-methyl-5-nitro-2-(trifluoromethyl)-2,3-dihydrochromen-4-one, IQ motif and SEC7 domain-containing protein 1, NONAETHYLENE GLYCOL | | Authors: | Nawrotek, A, Zeghouf, M, Cherfils, J. | | Deposit date: | 2018-02-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | PH-domain-binding inhibitors of nucleotide exchange factor BRAG2 disrupt Arf GTPase signaling.
Nat.Chem.Biol., 15, 2019
|
|
4JAK
 
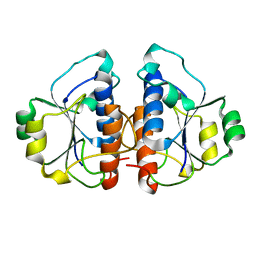 | |
6GB8
 
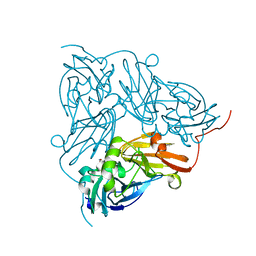 | | Copper nitrite reductase from Achromobacter cycloclastes: small cell polymorph dataset 1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
