7CW4
 
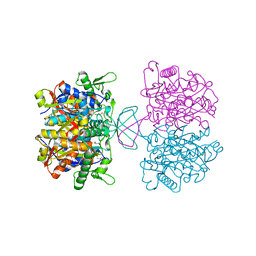 | | Acetyl-CoA acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Hong, J, Kim, K.J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of an acetyl-CoA acetyltransferase from PHB producing bacterium Bacillus cereus ATCC 14579.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7CW5
 
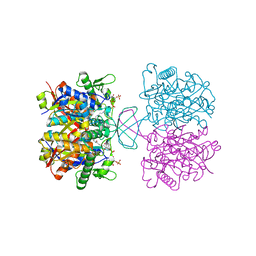 | | Acetyl-CoA acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A | | Authors: | Hong, J, Kim, K.J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an acetyl-CoA acetyltransferase from PHB producing bacterium Bacillus cereus ATCC 14579.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
8TUM
 
 | |
8TUX
 
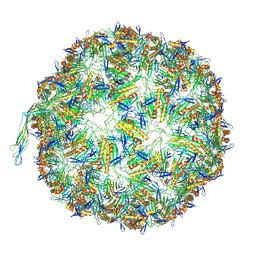 | |
8TUW
 
 | |
7WZ6
 
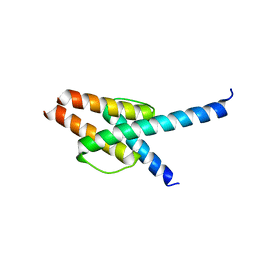 | | Crystal structure of MyoD-E47 | | Descriptor: | Isoform E47 of Transcription factor E2-alpha, Myoblast determination protein 1 | | Authors: | Zhong, J, Huang, Y, Ma, J. | | Deposit date: | 2022-02-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of the bHLH domains of MyoD-E47 heterodimer.
Biochem.Biophys.Res.Commun., 621, 2022
|
|
8YM7
 
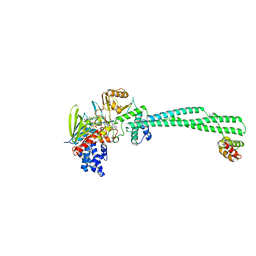 | | Crystal structure of Lysine Specific Demethylase 1 (LSD1) with JH-45 | | Descriptor: | 4-[5-(4-azanylpiperidin-1-yl)-8-(4-methylphenyl)pyrido[3,4-b]pyrazin-7-yl]-2-fluoranyl-benzenecarbonitrile, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, ... | | Authors: | Zhiyan, D, Danyan, C, Hong, J, Tongchao, L, Bing, X. | | Deposit date: | 2024-03-08 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of Novel LSD1 Inhibitors for the Treatment of Autosomal Dominant Polycystic Kidney Disease
To Be Published
|
|
3R22
 
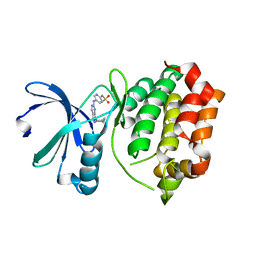 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesana, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R21
 
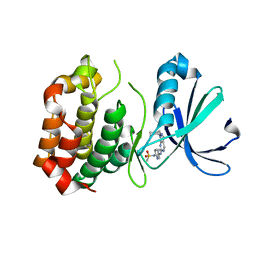 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | MAGNESIUM ION, N-(2-aminoethyl)-N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesena, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6NR0
 
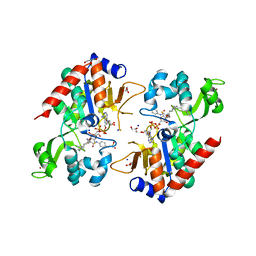 | |
8K9R
 
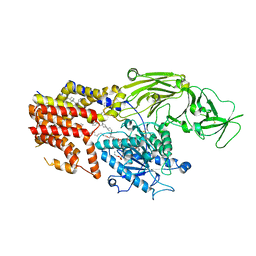 | | Cryo EM structure of the products-bound PGAP1(Bst1)-H443N from Chaetomium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
8K9T
 
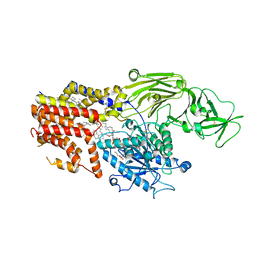 | | Cryo-EM structure of the products-bound PGAP1(Bst1)-S327A from Chaetonium thermophilum | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose, 2-azanylethyl [(2R,3S,4S,5S,6S)-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hydrogen phosphate, 2-azanylethyl [(2~{S},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl] hydrogen phosphate, ... | | Authors: | Li, T, Hong, J, Qu, Q, Li, D. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Molecular basis of the inositol deacylase PGAP1 involved in quality control of GPI-AP biogenesis.
Nat Commun, 15, 2024
|
|
1IL6
 
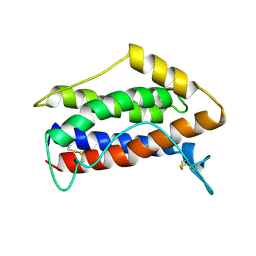 | | HUMAN INTERLEUKIN-6, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INTERLEUKIN-6 | | Authors: | Xu, G.Y, Yu, H.A, Hong, J, Stahl, M, Mcdonagh, T, Kay, L.E, Cumming, D.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of recombinant human interleukin-6.
J.Mol.Biol., 268, 1997
|
|
7TPR
 
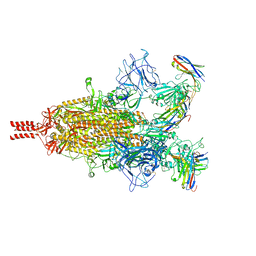 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | Descriptor: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | Authors: | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
3E9G
 
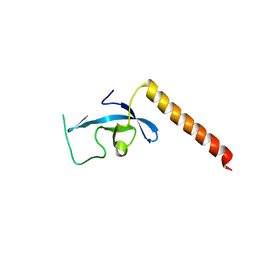 | | Crystal structure long-form (residue1-124) of Eaf3 chromo domain | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | Deposit date: | 2008-08-22 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
3JB8
 
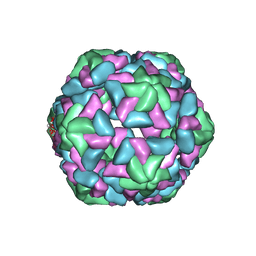 | | Insight into Three-dimensional structure of Maize Chlorotic Mottle Virus Revealed by Single Particle Analysis | | Descriptor: | Coat protein | | Authors: | Wang, C.Y, Zhang, Q.F, Gao, Y.Z, Zhou, X.P, Ji, G, Huang, X.J, Hong, J, Zhang, C.X. | | Deposit date: | 2015-08-04 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Insight into the three-dimensional structure of maize chlorotic mottle virus revealed by Cryo-EM single particle analysis.
Virology, 485, 2015
|
|
3E9F
 
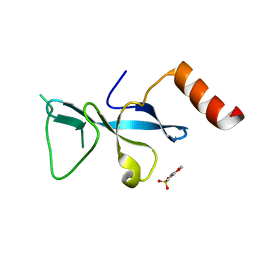 | | Crystal structure short-form (residue1-113) of Eaf3 chromo domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromatin modification-related protein EAF3 | | Authors: | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | Deposit date: | 2008-08-22 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
7YP1
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement) | | Descriptor: | 10E4 heavy chain, 10E4 light chain, EBV gH, ... | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YOY
 
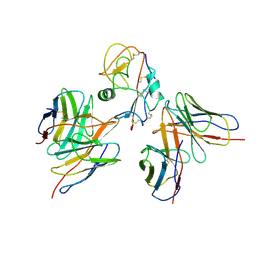 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement) | | Descriptor: | 3E8 heavy chain, 3E8 light chain, 5E3 heavy chain, ... | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YP2
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement) | | Descriptor: | 6H2 heavy chain, 6H2 light chain, Envelope glycoprotein H | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
6BUZ
 
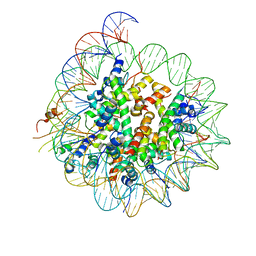 | | Cryo-EM structure of CENP-A nucleosome in complex with kinetochore protein CENP-N | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B, ... | | Authors: | Chittori, S, Hong, J, Kelly, A.E, Bai, Y, Subramaniam, S. | | Deposit date: | 2017-12-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N.
Science, 359, 2018
|
|
4J4Q
 
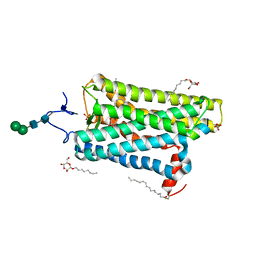 | | Crystal structure of active conformation of GPCR opsin stabilized by octylglucoside | | Descriptor: | ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, PALMITIC ACID, ... | | Authors: | Park, J.H, Morizumi, T, Li, Y, Hong, J.E, Pai, E.F, Hofmann, K.P, Choe, H.W, Ernst, O.P. | | Deposit date: | 2013-02-07 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Opsin, a structural model for olfactory receptors?
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2IL6
 
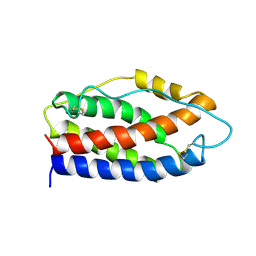 | | HUMAN INTERLEUKIN-6, NMR, 32 STRUCTURES | | Descriptor: | INTERLEUKIN-6 | | Authors: | Xu, G.Y, Yu, H.A, Hong, J, Stahl, M, Mcdonagh, T, Kay, L.E, Cumming, D.A. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of recombinant human interleukin-6.
J.Mol.Biol., 268, 1997
|
|
5CKR
 
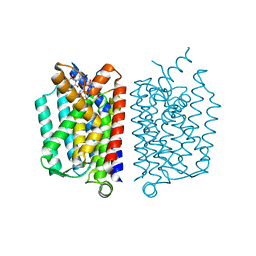 | | Crystal Structure of MraY in complex with Muraymycin D2 | | Descriptor: | Muraymycin D2, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Mashalidis, E.H, Tanino, T, Kim, M, Hong, J, Ichikawa, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into inhibition of lipid I production in bacterial cell wall synthesis.
Nature, 533, 2016
|
|
4J72
 
 | | Crystal Structure of polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Gillespie, R.A, Kwon, D.Y, Guan, Z, Zhou, P, Hong, J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis.
Science, 341, 2013
|
|
