7L1U
 
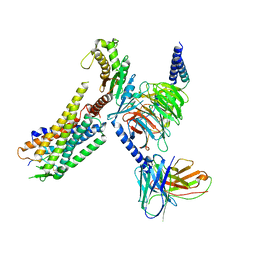 | | Orexin Receptor 2 (OX2R) in Complex with G Protein and Natural Peptide-Agonist Orexin B (OxB) | | 分子名称: | Engineered Guanine nucleotide-binding protein subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Hong, C, Byrne, N.J, Zamlynny, B, Tummala, S, Xiao, L, Shipman, J.M, Partridge, A.T, Minnick, C, Breslin, M.J, Rudd, M.T, Stachel, S.J, Rada, V.L, Kern, J.C, Armacost, K.A, Hollingsworth, S.A, O'Brien, J.A, Hall, D.L, McDonald, T.P, Strickland, C, Brooun, A, Soisson, S.M, Hollenstein, K. | | 登録日 | 2020-12-15 | | 公開日 | 2021-02-10 | | 最終更新日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of active-state orexin receptor 2 rationalize peptide and small-molecule agonist recognition and receptor activation.
Nat Commun, 12, 2021
|
|
7L1V
 
 | | Orexin Receptor 2 (OX2R) in Complex with G Protein and Small-Molecule Agonist Compound 1 | | 分子名称: | 4'-methoxy-N,N-dimethyl-3'-{[3-(2-{[2-(2H-1,2,3-triazol-2-yl)benzene-1-carbonyl]amino}ethyl)phenyl]sulfamoyl}[1,1'-biphenyl]-3-carboxamide, Engineered Guanine nucleotide-binding protein subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Hong, C, Byrne, N.J, Zamlynny, B, Tummala, S, Xiao, L, Shipman, J.M, Partridge, A.T, Minnick, C, Breslin, M.J, Rudd, M.T, Stachel, S.J, Rada, V.L, Kern, J.C, Armacost, K.A, Hollingsworth, S.A, O'Brien, J.A, Hall, D.L, McDonald, T.P, Strickland, C, Brooun, A, Soisson, S.M, Hollenstein, K. | | 登録日 | 2020-12-15 | | 公開日 | 2021-02-10 | | 最終更新日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of active-state orexin receptor 2 rationalize peptide and small-molecule agonist recognition and receptor activation.
Nat Commun, 12, 2021
|
|
5H4B
 
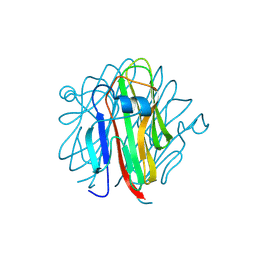 | | Crystal structure of Cbln4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-4 | | 著者 | Zhong, C, Shen, J, Zhang, H, Ding, J. | | 登録日 | 2016-10-31 | | 公開日 | 2017-09-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H49
 
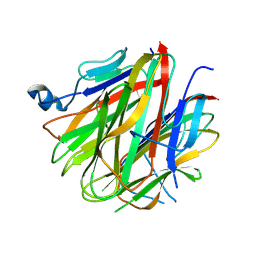 | | Crystal structure of Cbln1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | 著者 | Zhong, C, Shen, J, Zhang, H, Ding, J. | | 登録日 | 2016-10-31 | | 公開日 | 2017-09-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H4C
 
 | | Crystal structure of Cbln4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Cbln4 | | 著者 | Zhong, C, Shen, J, Zhang, H, Ding, J. | | 登録日 | 2016-10-31 | | 公開日 | 2017-09-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H48
 
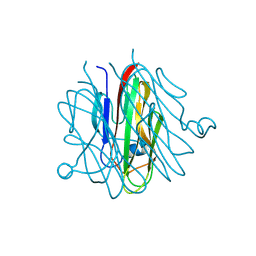 | | Crystal structure of Cbln1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | 著者 | Zhong, C, Shen, J, Zhang, H, Ding, J. | | 登録日 | 2016-10-31 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
1ITT
 
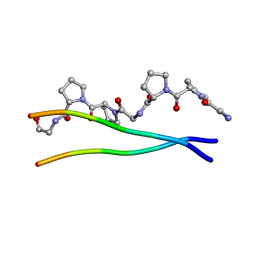 | | Average Crystal Structure of (Pro-Pro-Gly)9 at 1.0 angstroms Resolution | | 分子名称: | COLLAGEN TRIPLE HELIX | | 著者 | Hongo, C, Nagarajan, V, Noguchi, K, Kamitori, S, Okuyama, K, Tanaka, Y, Nishino, N. | | 登録日 | 2002-02-03 | | 公開日 | 2003-02-03 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Average Crystal Structure of (Pro-Pro-Gly)9 at 1.0 angstroms Resolution
Plym.J., 33, 2001
|
|
2CUO
 
 | | Collagen model peptide (PRO-PRO-GLY)9 | | 分子名称: | COLLAGEN MODEL PEPTIDE (PRO-PRO-GLY)9 | | 著者 | Hongo, C, Noguchi, K, Okuyama, K, Tanaka, Y, Nishino, N. | | 登録日 | 2005-05-27 | | 公開日 | 2005-06-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Repetitive interactions observed in the crystal structure of a collagen-model peptide, [(Pro-Pro-Gly)9]3
J.Biochem.(Tokyo), 138, 2005
|
|
5WTT
 
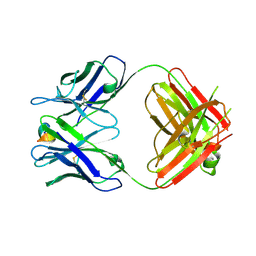 | | Structure of the 093G9 Fab in complex with the epitope peptide | | 分子名称: | Epitope peptide of Cyr61, Heavy chain of 093G9 Fab, Light chain of 093G9 Fab | | 著者 | Zhong, C, Hu, K, Shen, J, Ding, J. | | 登録日 | 2016-12-14 | | 公開日 | 2017-12-20 | | 最終更新日 | 2019-01-02 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular basis for the recognition of CCN1 by monoclonal antibody 093G9.
J. Mol. Recognit., 30, 2017
|
|
7V6F
 
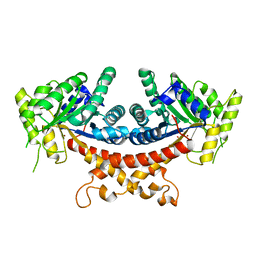 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase complexed with G3P | | 分子名称: | Fructose-bisphosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ZINC ION | | 著者 | Hongxuan, C, Huang, Y, Han, C, Chen, W, Ren, Y, Wan, J. | | 登録日 | 2021-08-20 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
6DWB
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome needle filament | | 分子名称: | Protein PrgI | | 著者 | Hu, J, Hong, C, Worrall, L.J, Vuckovic, M, Yu, Z, Strynadka, N.C.J. | | 登録日 | 2018-06-26 | | 公開日 | 2018-10-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6D04
 
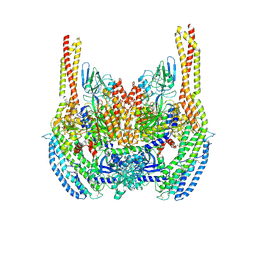 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; two molecules of parasite ligand, subclass 1. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | 登録日 | 2018-04-10 | | 公開日 | 2018-06-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
5JUY
 
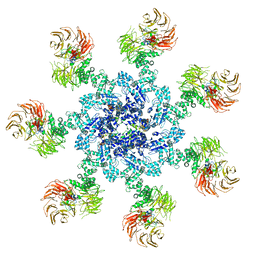 | | Active human apoptosome with procaspase-9 | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Caspase-9, ... | | 著者 | Cheng, T.C, Hong, C, Akey, I.V, Yuan, S, Akey, C.W. | | 登録日 | 2016-05-10 | | 公開日 | 2016-10-19 | | 最終更新日 | 2019-12-25 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | A near atomic structure of the active human apoptosome.
Elife, 5, 2016
|
|
5TCP
 
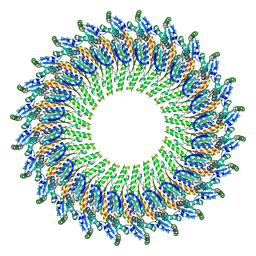 | | Near-atomic resolution cryo-EM structure of the periplasmic domains of PrgH and PrgK | | 分子名称: | Lipoprotein PrgK, Protein PrgH | | 著者 | Worrall, L.J, Hong, C, Vuckovic, M, Bergeron, J.R.C, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | 登録日 | 2016-09-15 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body.
Nature, 540, 2016
|
|
5TCQ
 
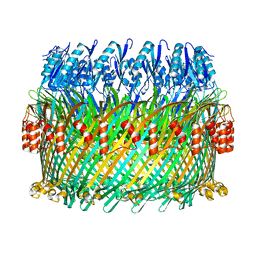 | | Near-atomic resolution cryo-EM structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG | | 分子名称: | Protein InvG | | 著者 | Worrall, L.J, Hong, C, Vuckovic, M, Bergeron, J.R.C, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | 登録日 | 2016-09-15 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body.
Nature, 540, 2016
|
|
5TCR
 
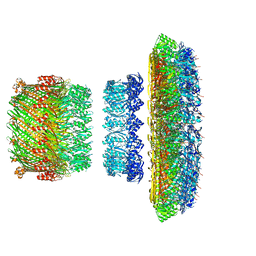 | | Atomic model of the Salmonella SPI-1 type III secretion injectisome basal body proteins InvG, PrgH, and PrgK | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH | | 著者 | Worrall, L.J, Hong, C, Vuckovic, M, Bergeron, J.R.C, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | 登録日 | 2016-09-15 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body.
Nature, 540, 2016
|
|
6NJP
 
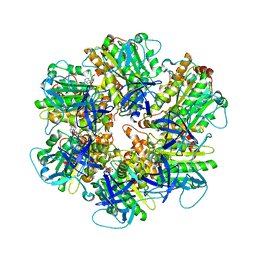 | | Structure of the assembled ATPase EscN in complex with its central stalk EscO from the enteropathogenic E. coli (EPEC) type III secretion system | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, EscO, ... | | 著者 | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | 登録日 | 2019-01-03 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
6NJO
 
 | | Structure of the assembled ATPase EscN from the enteropathogenic E. coli (EPEC) type III secretion system | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | 著者 | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | 登録日 | 2019-01-03 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
2DUV
 
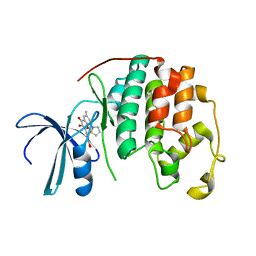 | | Structure of CDK2 with a 3-hydroxychromones | | 分子名称: | 2-(3,4-DIHYDROXYPHENYL)-8-(1,1-DIOXIDOISOTHIAZOLIDIN-2-YL)-3-HYDROXY-6-METHYL-4H-CHROMEN-4-ONE, Cell division protein kinase 2 | | 著者 | Kim, K.H, Lee, J, Park, T, Jeong, S, Hong, C. | | 登録日 | 2006-07-27 | | 公開日 | 2007-01-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | 3-Hydroxychromones as cyclin-dependent kinase inhibitors: synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5WC3
 
 | | SpoIIIAG | | 分子名称: | SpoIIIAG, Stage III sporulation engulfment assemblyprotein | | 著者 | Zeytuni, N, Hong, C, Worrall, L.J, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | 登録日 | 2017-06-29 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Near-atomic resolution cryoelectron microscopy structure of the 30-fold homooligomeric SpoIIIAG channel essential to spore formation in Bacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6D03
 
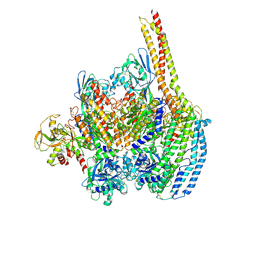 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; one molecule of parasite ligand. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(2-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | 登録日 | 2018-04-10 | | 公開日 | 2018-06-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6D05
 
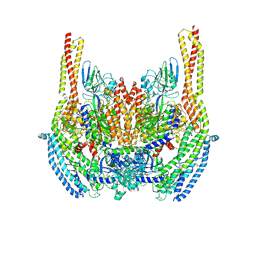 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; two molecules of parasite ligand, subclass 2. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | 登録日 | 2018-04-10 | | 公開日 | 2018-06-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
6B6H
 
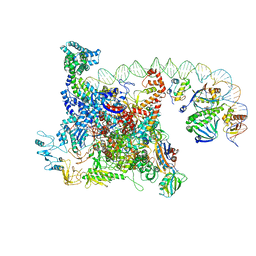 | | The cryo-EM structure of a bacterial class I transcription activation complex | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Liu, B, Hong, C, Huang, R, Yu, Z, Steitz, T.A. | | 登録日 | 2017-10-02 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis of bacterial transcription activation.
Science, 358, 2017
|
|
4KDF
 
 | |
4KDE
 
 | |
