1CTI
 
 | |
2CTI
 
 | |
3CTI
 
 | |
3MN7
 
 | | Structures of actin-bound WH2 domains of Spire and the implication for filament nucleation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Ducka, A.M, Sitar, T, Popowicz, G.M, Huber, R, Holak, T.A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6YCR
 
 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | FFIVIRDRVFR(CCS)G(NH2), Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Grudnik, P, Kuska, K, Holak, T.A, Dubin, G. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Macrocyclic Peptide Inhibitor of PD-1/PD-L1 Immune Checkpoint
Adv. Ther., 2020
|
|
4PYU
 
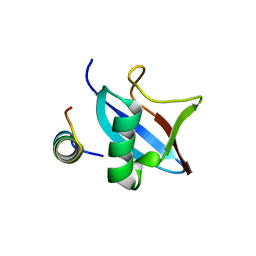 | | The conserved ubiquitin-like protein hub1 plays a critical role in splicing in human cells | | Descriptor: | U4/U6.U5 tri-snRNP-associated protein 1, Ubiquitin-like protein 5 | | Authors: | Ammon, T, Mishra, S.K, Kowalska, K, Popowicz, G.M, Holak, T.A, Jentsch, S. | | Deposit date: | 2014-03-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The conserved ubiquitin-like protein Hub1 plays a critical role in splicing in human cells.
J Mol Cell Biol, 6, 2014
|
|
5C3T
 
 | | PD-1 binding domain from human PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Dubin, G, Holak, T.A. | | Deposit date: | 2015-06-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1.
Structure, 23, 2015
|
|
3DAC
 
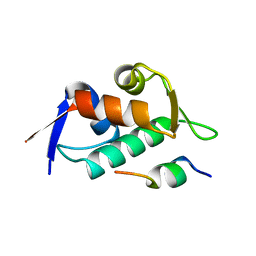 | |
3DAB
 
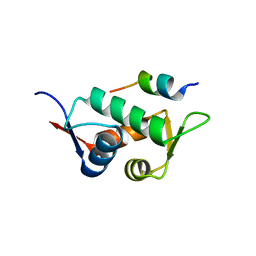 | |
5O4Y
 
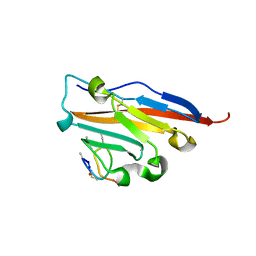 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | PHE-MAA-ASN-PRO-HIS-LEU-SER-TRP-SER-TRP-9KK-9KK-ARG-CCS-GLY-NH2, Programmed cell death 1 ligand 1 | | Authors: | Magiera, K, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bioactive Macrocyclic Inhibitors of the PD-1/PD-L1 Immune Checkpoint.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5OAI
 
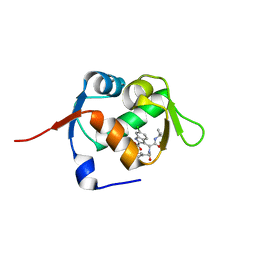 | | Structure of MDM2 with low molecular weight inhibitor | | Descriptor: | 3-[(1~{R})-2-(~{tert}-butylamino)-1-[methanoyl-[[3,4,5-tris(fluoranyl)phenyl]methyl]amino]-2-oxidanylidene-ethyl]-6-chloranyl-1~{H}-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Neochoritis, C.G, Grudnik, P, Dubin, G, Domling, A, Holak, T.A. | | Deposit date: | 2017-06-22 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A fluorinated indole-based MDM2 antagonist selectively inhibits the growth of p53wtosteosarcoma cells.
Febs J., 286, 2019
|
|
5O45
 
 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | PHE-MEA-9KK-SAR-ASP-VAL-MEA-TYR-SAR-TRP-TYR-LEU-CCS-GLY-NH2, Programmed cell death 1 ligand 1 | | Authors: | Magiera, K, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-05-26 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Bioactive Macrocyclic Inhibitors of the PD-1/PD-L1 Immune Checkpoint.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4MDQ
 
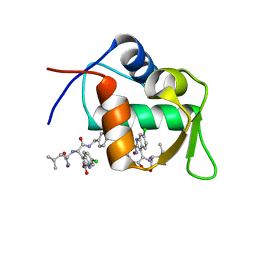 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-[(1R)-2-(benzylamino)-1-{[(2S)-1-(hydroxyamino)-4-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl]-6-chloro-N-hydroxy-1H-indole-2-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Bista, M, Popowicz, G, Holak, T.A. | | Deposit date: | 2013-08-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
4MDN
 
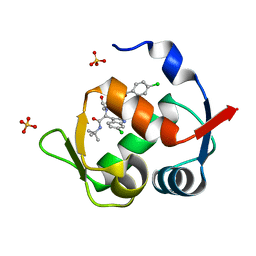 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-{(1S)-2-(tert-butylamino)-1-[{4-[(4-chlorobenzyl)oxy]benzyl}(formyl)amino]-2-oxoethyl}-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Bista, M, Popowicz, G, Holak, T.A. | | Deposit date: | 2013-08-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
3FDO
 
 | |
3G03
 
 | |
3V3B
 
 | | Structure of the Stapled p53 Peptide Bound to Mdm2 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, SAH-p53-8 stapled-peptide | | Authors: | Baek, S, Kutchukian, P.S, Verdine, G.L, Huber, R, Holak, T.A, Ki Won, L, Popowicz, G.M. | | Deposit date: | 2011-12-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the stapled p53 peptide bound to Mdm2.
J.Am.Chem.Soc., 134, 2012
|
|
3V3V
 
 | | Structural and functional analysis of quercetagetin, a natural JNK1 inhibitor | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, C-Jun-amino-terminal kinase-interacting protein 1, CHLORIDE ION, ... | | Authors: | Baek, S, Kang, N.J, Popowicz, G.M, Arciniega, M, Jung, S.K, Byun, S, Song, N.R, Heo, Y.S, Kim, B.Y, Lee, H.J, Holak, T.A, Augustin, M, Bode, A.M, Huber, R, Dong, Z, Lee, K.W. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of the Natural JNK1 Inhibitor Quercetagetin.
J.Mol.Biol., 425, 2013
|
|
1DNY
 
 | | SOLUTION STRUCTURE OF PCP, A PROTOTYPE FOR THE PEPTIDYL CARRIER DOMAINS OF MODULAR PEPTIDE SYNTHETASES | | Descriptor: | NON-RIBOSOMAL PEPTIDE SYNTHETASE PEPTIDYL CARRIER PROTEIN | | Authors: | Weber, T, Baumgartner, R, Renner, C, Marahiel, M.A, Holak, T.A. | | Deposit date: | 1999-12-17 | | Release date: | 2000-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PCP, a prototype for the peptidyl carrier domains of modular peptide synthetases.
Structure Fold.Des., 8, 2000
|
|
4ZQK
 
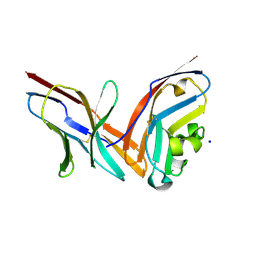 | | Structure of the complex of human programmed death-1 (PD-1) and its ligand PD-L1. | | Descriptor: | Programmed cell death 1 ligand 1, Programmed cell death protein 1, SODIUM ION | | Authors: | Zak, K.M, Dubin, G, Holak, T.A. | | Deposit date: | 2015-05-10 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1.
Structure, 23, 2015
|
|
1DTV
 
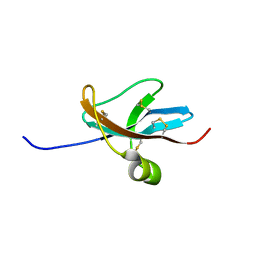 | | NMR STRUCTURE OF THE LEECH CARBOXYPEPTIDASE INHIBITOR (LCI) | | Descriptor: | CARBOXYPEPTIDASE INHIBITOR | | Authors: | Reverter, D, Fernandez-Catalan, C, Bode, W, Holak, T.A, Aviles, F.X. | | Deposit date: | 2000-01-13 | | Release date: | 2000-07-19 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of a novel leech carboxypeptidase inhibitor determined free in solution and in complex with human carboxypeptidase A2.
Nat.Struct.Biol., 7, 2000
|
|
1DTD
 
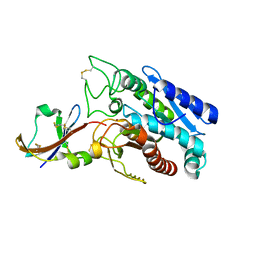 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN THE LEECH CARBOXYPEPTIDASE INHIBITOR AND THE HUMAN CARBOXYPEPTIDASE A2 (LCI-CPA2) | | Descriptor: | CARBOXYPEPTIDASE A2, GLUTAMIC ACID, METALLOCARBOXYPEPTIDASE INHIBITOR, ... | | Authors: | Reverter, D, Fernandez-Catalan, C, Bode, W, Holak, T.A, Aviles, F.X. | | Deposit date: | 2000-01-12 | | Release date: | 2000-07-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a novel leech carboxypeptidase inhibitor determined free in solution and in complex with human carboxypeptidase A2.
Nat.Struct.Biol., 7, 2000
|
|
5N2F
 
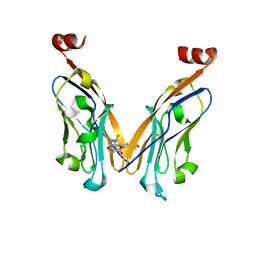 | | Structure of PD-L1/small-molecule inhibitor complex | | Descriptor: | 4-[[4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-2,5-bis(fluoranyl)phenyl]methylamino]-3-oxidanylidene-butanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
5N86
 
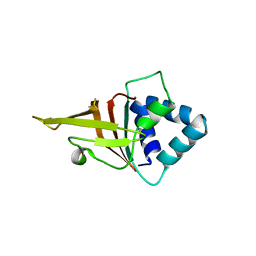 | | Crystal structure of FAS1 domain of hyaluronic acid receptor stabilin-2 | | Descriptor: | Stabilin-2 | | Authors: | Twarda-Clapa, A, Labuzek, B, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Crystal structure of the FAS1 domain of the hyaluronic acid receptor stabilin-2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4ZFI
 
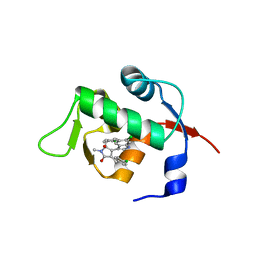 | | Structure of Mdm2 with low molecular weight inhibitor | | Descriptor: | (5S)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxy-1-methyl-1,5-dihydro-2H-pyrrol-2-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Zak, K.M, Twarda-Clapa, A, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2015-04-21 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
