2CTI
 
 | |
1CTI
 
 | |
2ABZ
 
 | | Crystal structure of C19A/C43A mutant of leech carboxypeptidase inhibitor in complex with bovine carboxypeptidase A | | Descriptor: | Carboxypeptidase A1, Metallocarboxypeptidase inhibitor, ZINC ION | | Authors: | Arolas, J.L, Popowicz, G.M, Bronsoms, S, Aviles, F.X, Huber, R, Holak, T.A, Ventura, S. | | Deposit date: | 2005-07-18 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Study of a major intermediate in the oxidative folding of leech carboxypeptidase inhibitor: contribution of the fourth disulfide bond
J.Mol.Biol., 352, 2005
|
|
5J7G
 
 | | Structure of MDM2 with low molecular weight inhibitor with aliphatic linker. | | Descriptor: | 4-({6-[(6-chloro-3-{1-[(4-chlorophenyl)methyl]-4-(4-fluorophenyl)-1H-imidazol-5-yl}-1H-indole-2-carbonyl)oxy]hexyl}amino)-4-oxobutanoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Kubica, K, Guzik, K, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,4,5-Trisubstituted Imidazole-Based p53-MDM2/MDMX Antagonists with Aliphatic Linkers for Conjugation with Biological Carriers.
J. Med. Chem., 60, 2017
|
|
8P1O
 
 | | Solubilizer tag effect on PD-L1/inhibitor binding properties for m-terphenyl derivatives | | Descriptor: | (3~{R})-1-[[4-[2-chloranyl-3-(2,3-dihydro-1,4-benzodioxin-6-yl)phenyl]-2-methoxy-phenyl]methyl]-~{N}-(2-hydroxyethyl)pyrrolidine-3-carboxamide, CHLORIDE ION, Programmed cell death 1 ligand 1, ... | | Authors: | Plewka, J, Magiera-Mularz, K, Surmiak, E, Kalinowska-Tluscik, J, Holak, T.A. | | Deposit date: | 2023-05-12 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Solubilizer Tag Effect on PD-L1/Inhibitor Binding Properties for m -Terphenyl Derivatives.
Acs Med.Chem.Lett., 15, 2024
|
|
4PYU
 
 | | The conserved ubiquitin-like protein hub1 plays a critical role in splicing in human cells | | Descriptor: | U4/U6.U5 tri-snRNP-associated protein 1, Ubiquitin-like protein 5 | | Authors: | Ammon, T, Mishra, S.K, Kowalska, K, Popowicz, G.M, Holak, T.A, Jentsch, S. | | Deposit date: | 2014-03-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The conserved ubiquitin-like protein Hub1 plays a critical role in splicing in human cells.
J Mol Cell Biol, 6, 2014
|
|
5C3T
 
 | | PD-1 binding domain from human PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Dubin, G, Holak, T.A. | | Deposit date: | 2015-06-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1.
Structure, 23, 2015
|
|
5J7F
 
 | | Structure of MDM2 with low molecular weight inhibitor with aliphatic linker. | | Descriptor: | 4-({6-[(6-chloro-3-{1-[(4-chlorophenyl)methyl]-4-(4-fluorophenyl)-1H-imidazol-5-yl}-1H-indole-2-carbonyl)oxy]hexyl}amino)-4-oxobutanoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Kubica, K, Guzik, K, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4,5-Trisubstituted Imidazole-Based p53-MDM2/MDMX Antagonists with Aliphatic Linkers for Conjugation with Biological Carriers.
J. Med. Chem., 60, 2017
|
|
6YCR
 
 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | FFIVIRDRVFR(CCS)G(NH2), Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Grudnik, P, Kuska, K, Holak, T.A, Dubin, G. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Macrocyclic Peptide Inhibitor of PD-1/PD-L1 Immune Checkpoint
Adv. Ther., 2020
|
|
1I16
 
 | | STRUCTURE OF INTERLEUKIN 16: IMPLICATIONS FOR FUNCTION, NMR, 20 STRUCTURES | | Descriptor: | INTERLEUKIN 16 | | Authors: | Muehlhahn, P, Zweckstetter, M, Georgescu, J, Ciosto, C, Renner, C, Lanzendoerfer, M, Lang, K, Ambrosius, D, Baier, M, Kurth, R, Holak, T.A. | | Deposit date: | 1998-05-20 | | Release date: | 1999-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of interleukin 16 resembles a PDZ domain with an occluded peptide binding site.
Nat.Struct.Biol., 5, 1998
|
|
1HJ0
 
 | | Thymosin beta9 | | Descriptor: | THYMOSIN BETA9 | | Authors: | Stoll, R, Voelter, W, Holak, T.A. | | Deposit date: | 2001-01-05 | | Release date: | 2002-01-04 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Conformation of Thymosin Beta9 in Water/Fluoroalcohol Solution Determined by NMR Spectroscopy
Biopolymers, 41, 1997
|
|
5J8O
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2R)-1-({3-bromo-4-[(2-methyl[1,1'-biphenyl]-3-yl)methoxy]phenyl}methyl)piperidine-2-carboxylic acid, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Guzik, K, Zieba, B.J, Musielak, B, Doemling, P, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1).
Oncotarget, 7, 2016
|
|
5J89
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-{2-[({2-methoxy-6-[(2-methyl[1,1'-biphenyl]-3-yl)methoxy]pyridin-3-yl}methyl)amino]ethyl}acetamide, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Guzik, K, Zieba, B.J, Musielak, B, Doemling, P, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-07 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1).
Oncotarget, 7, 2016
|
|
2Z5S
 
 | | Molecular basis for the inhibition of p53 by Mdmx | | Descriptor: | Cellular tumor antigen p53, Mdm4 protein | | Authors: | Popowicz, G.M, Czarna, A, Rothweiler, U, Szwagierczak, A, Holak, T.A. | | Deposit date: | 2007-07-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of p53 by Mdmx.
Cell Cycle, 6, 2007
|
|
2Z5T
 
 | | Molecular basis for the inhibition of p53 by Mdmx | | Descriptor: | Cellular tumor antigen p53, Mdm4 protein | | Authors: | Popowicz, G.M, Czarna, A, Rothweiler, U, Szwagierczak, A, Holak, T.A. | | Deposit date: | 2007-07-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of p53 by Mdmx.
Cell Cycle, 6, 2007
|
|
3CTI
 
 | |
3PLV
 
 | | Structure of Hub-1 protein in complex with Snu66 peptide (HINDII) | | Descriptor: | 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Ubiquitin-like modifier HUB1 | | Authors: | Mishra, S.K, Ammon, T, Popowicz, G.M, Krajewski, M, Nagel, R.J, Ares, M, Holak, T.A, Jentsch, S. | | Deposit date: | 2010-11-15 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing.
Nature, 474, 2011
|
|
3G03
 
 | |
3PLU
 
 | | Structure of Hub-1 protein in complex with Snu66 peptide (HINDI) | | Descriptor: | 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Ubiquitin-like modifier HUB1 | | Authors: | Mishra, S.K, Ammon, T, Popowicz, G.M, Krajewski, M, Nagel, R.J, Ares, M, Holak, T.A, Jentsch, S. | | Deposit date: | 2010-11-15 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing.
Nature, 474, 2011
|
|
3RMP
 
 | | Structural basis for the recognition of attP substrates by P4-like integrases | | Descriptor: | 5'-D(*TP*AP*AP*TP*GP*AP*CP*CP*AP*CP*CP*AP*AP*TP*A)-3', 5'-D(*TP*AP*TP*TP*GP*GP*TP*GP*GP*TP*CP*AP*TP*TP*A)-3', CP4-like integrase | | Authors: | Szwagierczak, A, Popowicz, G.M, Holak, T.A, Rakin, A, Antonenka, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for the recognition of attP substrates by P4-like integrases
To be Published
|
|
3JTZ
 
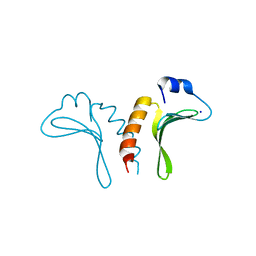 | | Structure of the arm-type binding domain of HPI integrase | | Descriptor: | Integrase, SODIUM ION | | Authors: | Szwagierczak, A, Antonenka, U, Popowicz, G.M, Sitar, T, Holak, T.A, Rakin, A. | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the arm-type binding domains of HPI and HAI7 integrases
J.Biol.Chem., 284, 2009
|
|
3JU0
 
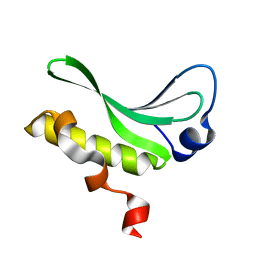 | | Structure of the arm-type binding domain of HAI7 integrase | | Descriptor: | Phage integrase | | Authors: | Szwagierczak, A, Antonenka, U, Popowicz, G.M, Sitar, T, Holak, T.A, Rakin, A. | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the arm-type binding domains of HPI and HAI7 integrases
J.Biol.Chem., 284, 2009
|
|
6RPG
 
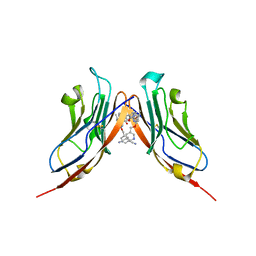 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with inhibitor | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[2-[[4-[[3-[3-[[4-[(2-acetamidoethylamino)methyl]-5-[(5-cyanopyridin-3-yl)methoxy]-2-methyl-phenoxy]methyl]-2-methyl-phenyl]-2-methyl-phenyl]methoxy]-2-[(5-cyanopyridin-3-yl)methoxy]-5-methyl-phenyl]methylamino]ethyl]ethanamide | | Authors: | Magiera-Mularz, K, Basu, S, Yang, J, Xu, B, Skalniak, L, Musielak, B, Kholodovych, V, Holak, T.A, Hu, L. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, Evaluation, and Structural Studies ofC2-Symmetric Small Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6SRU
 
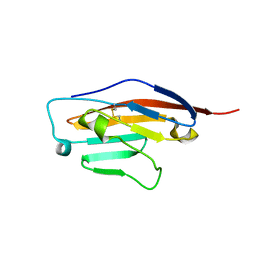 | | Structure of Ig-like V-type domian of mouse Programmed cell death 1 ligand 1 (PD-L1) | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Sala, D, Grudnik, P, Holak, T.A. | | Deposit date: | 2019-09-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Human and mouse PD-L1: similar molecular structure, but different druggability profiles.
Iscience, 24, 2021
|
|
6R3K
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2~{S},4~{R})-1-[[5-chloranyl-2-[(3-cyanophenyl)methoxy]-4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Skalniak, L, Dubin, G, Holak, T.A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Terphenyl-Based Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
