6IRS
 
 | | human LAT1-4F2hc complex incubated with JPH203 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Yan, R.H, Zhao, X, Lei, J.L, Zhou, Q. | | Deposit date: | 2018-11-14 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex.
Nature, 568, 2019
|
|
8WTI
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 20j | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Zeng, R, Zhao, X, Yang, S.Y, Lei, J. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of alpha-Ketoamide inhibitors of SARS-CoV-2 main protease derived from quaternized P1 groups.
Bioorg.Chem., 143, 2024
|
|
3HTK
 
 | | Crystal structure of Mms21 and Smc5 complex | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ZINC ION | | Authors: | Duan, X, Sarangi, P, Liu, X, Rangi, G.K, Zhao, X, Ye, H. | | Deposit date: | 2009-06-11 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex.
Mol.Cell, 35, 2009
|
|
2PZF
 
 | | Minimal human CFTR first nucleotide binding domain as a head-to-tail dimer with delta F508 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Atwell, S, Conners, K, Emtage, S, Gheyi, T, Glenn, N.R, Hendle, J, Lewis, H.A, Lu, F, Rodgers, L.A, Romero, R, Sauder, J.M, Smith, D, Tien, H, Wasserman, S.R, Zhao, X. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant.
Protein Eng.Des.Sel., 23, 2010
|
|
3PL9
 
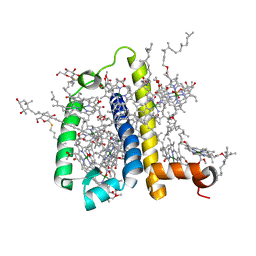 | | Crystal structure of spinach minor light-harvesting complex CP29 at 2.80 angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Li, M, Wan, T, Wang, L.F, Jia, C.J, Hou, Z.Q, Zhao, X.L, Zhang, J.P, Chang, W.R. | | Deposit date: | 2010-11-14 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into energy regulation of light-harvesting complex CP29 from spinach.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8V0C
 
 | | Structure of TDP1 catalytic domain complexed with compound IB06 | | Descriptor: | (8M)-8-(2-{[2-(fluorosulfonyl)ethyl]amino}phenyl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke Jr, T.R. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of TDP1 complexed with inhibitors
To Be Published
|
|
2PZG
 
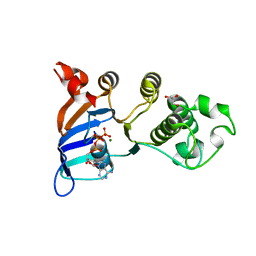 | | Minimal human CFTR first nucleotide binding domain as a monomer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, GLYCEROL, ... | | Authors: | Atwell, S, Conners, K, Emtage, S, Gheyi, T, Glenn, N.R, Hendle, J, Lewis, H.A, Lu, F, Rodgers, L.A, Romero, R, Sauder, J.M, Smith, D, Tien, H, Wasserman, S.R, Zhao, X. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant.
Protein Eng.Des.Sel., 23, 2010
|
|
2PZE
 
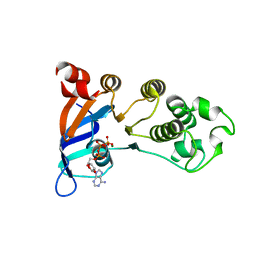 | | Minimal human CFTR first nucleotide binding domain as a head-to-tail dimer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Atwell, S, Conners, K, Emtage, S, Gheyi, T, Glenn, N.R, Hendle, J, Lewis, H.A, Lu, F, Rodgers, L.A, Romero, R, Sauder, J.M, Smith, D, Tien, H, Wasserman, S.R, Zhao, X. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant.
Protein Eng.Des.Sel., 23, 2010
|
|
5XZ3
 
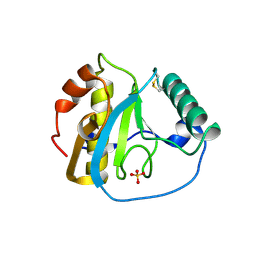 | | The X-ray structure of Apis mellifera PGRP-SA | | Descriptor: | Peptidoglycan-recognition protein, SULFATE ION | | Authors: | Liu, Y.J, Huang, J.X, Zhao, X.M, An, J.D. | | Deposit date: | 2017-07-11 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of peptidoglycan recognition protein SA in Apis mellifera (Hymenoptera: Apidae).
Protein Sci., 27, 2018
|
|
7SDE
 
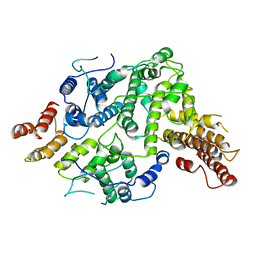 | | Cryo-EM structure of Nse5/6 heterodimer | | Descriptor: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | Authors: | Yu, Y, Patel, D.J, Zhao, X.L. | | Deposit date: | 2021-09-29 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The cryo-EM structure of Nse5/6 complex with the C terminal part of Nse5
To Be Published
|
|
8WY7
 
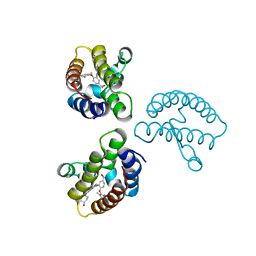 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 22 | | Descriptor: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 4 | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
7UFZ
 
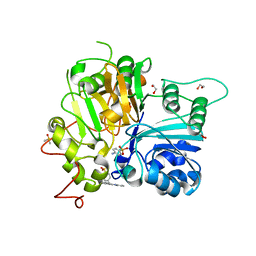 | | Crystal structure of TDP1 complexed with compound XZ768 | | Descriptor: | (4-{[(4S)-2-phenylimidazo[1,2-a]pyridin-3-yl]amino}phenyl)phosphonic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Wang, W, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | Deposit date: | 2022-03-23 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.559 Å) | | Cite: | Phosphonic acid-containing inhibitors of tyrosyl-DNA phosphodiesterase 1.
Front Chem, 10, 2022
|
|
7UFY
 
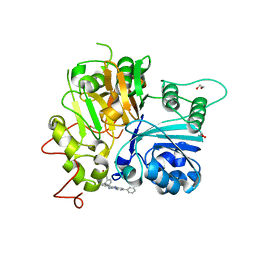 | | Crystal structure of TDP1 complexed with compound XZ766 | | Descriptor: | 1,2-ETHANEDIOL, Tyrosyl-DNA phosphodiesterase 1, [(4-{[(4S)-2,7-diphenylimidazo[1,2-a]pyridin-3-yl]amino}phenyl)methyl]phosphonic acid | | Authors: | Lountos, G.T, Zhao, X.Z, Wang, W, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | Deposit date: | 2022-03-23 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Phosphonic acid-containing inhibitors of tyrosyl-DNA phosphodiesterase 1.
Front Chem, 10, 2022
|
|
8WY3
 
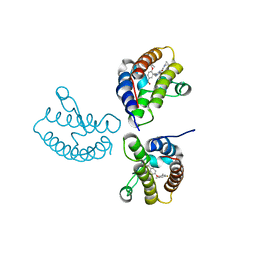 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 21 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-cyclopropyl-2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]ethanamide | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8WYG
 
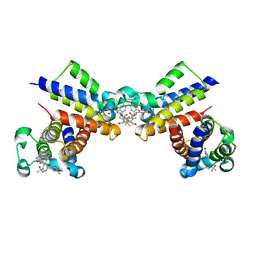 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor 22 | | Descriptor: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 2 | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8WXY
 
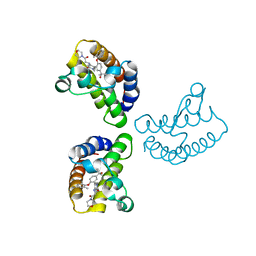 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 23 | | Descriptor: | 5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-4-[(2-morpholin-4-yl-2-oxidanylidene-ethyl)amino]pyridin-2-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
6CAA
 
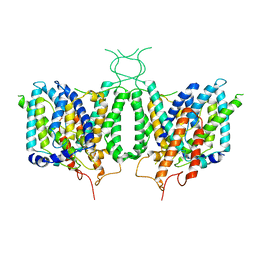 | | CryoEM structure of human SLC4A4 sodium-coupled acid-base transporter NBCe1 | | Descriptor: | Electrogenic sodium bicarbonate cotransporter 1 | | Authors: | Huynh, K.W, Jiang, J, Abuladze, N, Tsirulnikov, K, Kao, L, Shao, X, Newman, D, Azimov, R, Pushkin, A, Zhou, Z.H, Kurtz, I. | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM structure of the human SLC4A4 sodium-coupled acid-base transporter NBCe1.
Nat Commun, 9, 2018
|
|
8WQQ
 
 | |
7C9V
 
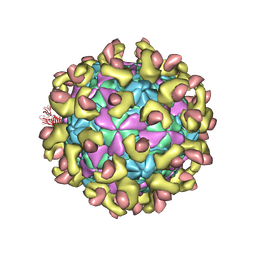 | | E30 F-particle in complex with FcRn | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, MYRISTIC ACID, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
1XKO
 
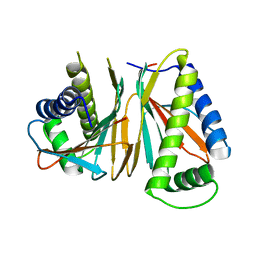 | | Structure of Thermotoga maritima CheX | | Descriptor: | CHEMOTAXIS protein cheX | | Authors: | Park, S.Y, Chao, X, Gonzalez-Bonet, G, Beel, B.D, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-09-29 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and Function of an Unusual Family of Protein Phosphatases; The Bacterial Chemotaxis Proteins CheC and CheX.
Mol.Cell, 16, 2004
|
|
7C9S
 
 | | Echovirus 30 F-particle | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Wang, K, Sun, Y, Zhu, L, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
5XZ4
 
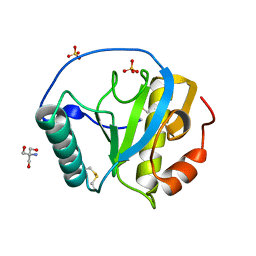 | | The X-tay structure of Bumblebee PGRP-SA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bumblebee peptidoglycan recognition protein SA, SULFATE ION | | Authors: | Liu, Y.J, Huang, J.X, Zhao, X.M, An, J.D. | | Deposit date: | 2017-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural Insights into the Preferential Binding of PGRP-SAs from Bumblebees and Honeybees to Dap-Type Peptidoglycans Rather than Lys-Type Peptidoglycans.
J Immunol., 202, 2019
|
|
7C9U
 
 | | Echovirus 30 E-particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9W
 
 | | E30 F-particle in complex with CD55 | | Descriptor: | Complement decay-accelerating factor, MYRISTIC ACID, SPHINGOSINE, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9T
 
 | | Echovirus 30 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
