3J05
 
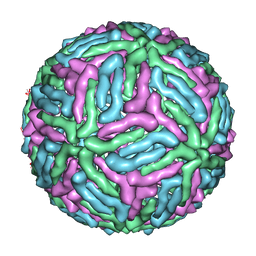 | | Three-dimensional structure of Dengue virus serotype 1 complexed with HMAb 14c10 Fab | | Descriptor: | envelope protein | | Authors: | Teoh, E.P, Kukkaro, P, Teo, E.W, Lim, A, Tan, T.T, Shi, P.Y, Yip, A, Schul, W, Leo, Y.S, Chan, S.H, Smith, K.G.C, Ooi, E.E, Kemeny, D.M, Ng, G, Ng, M.L, Alonso, S, Fisher, D, Hanson, B, Lok, S.M, MacAry, P.A. | | Deposit date: | 2011-04-01 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | The structural basis for serotype-specific neutralization of dengue virus by a human antibody.
Sci Transl Med, 4, 2012
|
|
3IWM
 
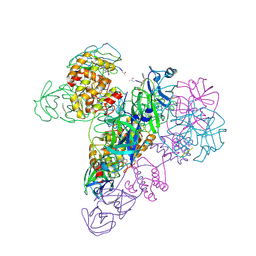 | | The octameric SARS-CoV main protease | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhong, N, Zhang, S, Xue, F, Lou, Z, Rao, Z, Xia, B. | | Deposit date: | 2009-09-02 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease
Protein Cell, 1, 2010
|
|
8EXX
 
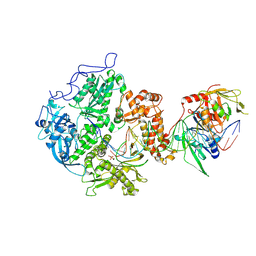 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and foscarnet (pre-translocation state) | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2022-10-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 2024
|
|
7WF3
 
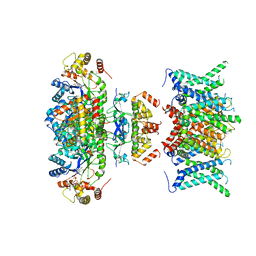 | | Composite map of human Kv1.3 channel in apo state with beta subunits | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, ... | | Authors: | Tyagi, A, Ahmed, T, Jian, S, Bajaj, S, Ong, S.T, Goay, S.S.M, Zhao, Y, Vorobyov, I, Tian, C, Chandy, K.G, Bhushan, S. | | Deposit date: | 2021-12-25 | | Release date: | 2022-02-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WF4
 
 | | Composite map of human Kv1.3 channel in dalazatide-bound state with beta subunits | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, ... | | Authors: | Tyagi, A, Ahmed, T, Jian, S, Bajaj, S, Ong, S.T, Goay, S.S.M, Zhao, Y, Vorobyov, I, Tian, C, Chandy, K.G, Bhushan, S. | | Deposit date: | 2021-12-25 | | Release date: | 2022-02-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7C7Q
 
 | | Cryo-EM structure of the baclofen/BHFF-bound human GABA(B) receptor in active state | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
5IV8
 
 | | The LPS Transporter LptDE from Klebsiella pneumoniae, core complex | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LPS biosynthesis protein, LPS-assembly lipoprotein LptE | | Authors: | Botos, I, McCarthy, J.G, Buchanan, S.K. | | Deposit date: | 2016-03-20 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.938 Å) | | Cite: | Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens.
Structure, 24, 2016
|
|
8TNN
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with gp42 antibody A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A10 heavy chain, A10 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
4QFJ
 
 | | The crystal structure of rat angiogenin-heparin complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Angiogenin, ... | | Authors: | Yeo, K.J, Hwang, E, Min, K.M, Hwang, K.Y, Jeon, Y.H, Chang, S.I, Cheong, H.K. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | The crystal structure of rat angiogenin-heparin complex
To be Published
|
|
8TNT
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with antibodies F-2-1 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 heavy chain, 769C2 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
8TOO
 
 | | Crystal structure of Epstein-Barr virus gp42 in complex with antibody 4C12 | | Descriptor: | 4C12 heavy chain, 4C12 light chain, Glycoprotein 42 | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
5IVA
 
 | |
8SW4
 
 | | BG505 GT1.1 SOSIP in complex with NHP Fabs 21N13, 21M20 and RM20A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 21M20 heavy chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Zhang, S, Ward, A.B. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-29 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Germline-targeting HIV vaccination induces neutralizing antibodies to the CD4 binding site.
Sci Immunol, 9, 2024
|
|
5TZ8
 
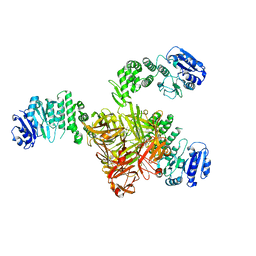 | | Crystal structure of S. aureus TarS | | Descriptor: | Glycosyl transferase | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
8UK9
 
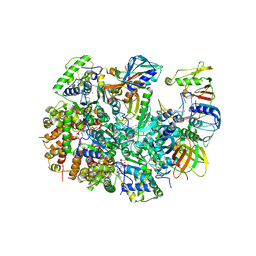 | | Structure of T4 Bacteriophage clamp loader mutant D110C bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, DNA primer, ... | | Authors: | Marcus, K, Ghaffari-Kashani, S, Gee, C.L. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5TZE
 
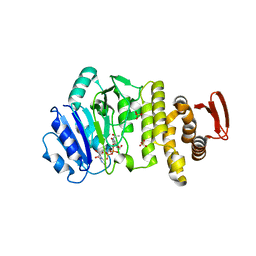 | | Crystal structure of S. aureus TarS in complex with UDP-GlcNAc | | Descriptor: | Glycosyl transferase, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
6EJZ
 
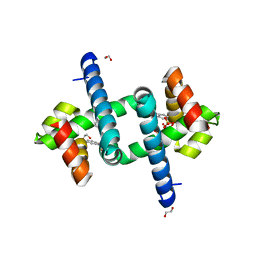 | | Tryptophan Repressor TrpR from E.coli variant S88Y with Indole-3-acetic acid as ligand | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, SULFATE ION, ... | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-09-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
5U02
 
 | | Crystal structure of S. aureus TarS 217-571 | | Descriptor: | Glycosyl transferase, IMIDAZOLE | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-22 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
7E2Q
 
 | | Crystal structure of Mycoplasma pneumoniae Enolase | | Descriptor: | Enolase, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Xiong, Q, Shao, G, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
6KZE
 
 | | The crystal structue of PDE10A complexed with 4d | | Descriptor: | 8-[(E)-2-[5-methyl-1-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]benzimidazol-2-yl]ethenyl]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, Y, Zhang, S, Zhou, Q, Huang, Y.-Y, Guo, L, Luo, H.-B. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.50003481 Å) | | Cite: | Novel Potent and Highly Selective Benzoimidazole-based Phosphodiesterase 10 Inhibitors with Improved Solubility and Pharmacokinetic Properties for the Treatment of Pulmonary Arterial Hypertension
To Be Published
|
|
5SV0
 
 | | Structure of the ExbB/ExbD complex from E. coli at pH 7.0 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Biopolymer transport protein ExbB, CALCIUM ION | | Authors: | Celia, H, Botos, I, Lloubes, R, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2016-08-04 | | Release date: | 2016-09-28 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into the role of the Ton complex in energy transduction.
Nature, 538, 2016
|
|
6EZ8
 
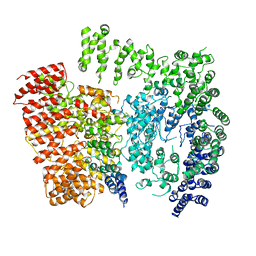 | | Human Huntingtin-HAP40 complex structure | | Descriptor: | Factor VIII intron 22 protein, Huntingtin | | Authors: | Guo, Q, Bin, H, Cheng, J, Pfeifer, G, Baumeister, W, Fernandez-Busnadiego, R, Kochanek, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-02-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-electron microscopy structure of huntingtin.
Nature, 555, 2018
|
|
3HQ2
 
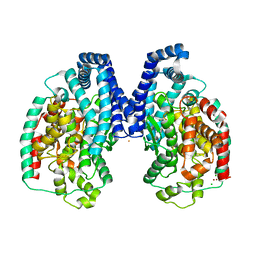 | | BsuCP Crystal Structure | | Descriptor: | Bacillus subtilis M32 carboxypeptidase, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Lee, M.M, Isaza, C.E, White, J.D, Chen, R.P.-Y, Liang, G.F.-C, He, H.T.-F, Chan, S.I, Chan, M.K. | | Deposit date: | 2009-06-05 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into the substrate length restriction of M32 carboxypeptidases: Characterization of two distinct subfamilies.
Proteins, 77, 2009
|
|
5DJA
 
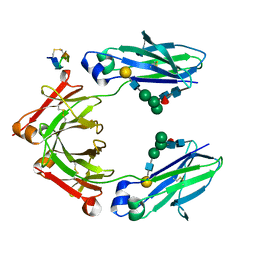 | | Fc Heterodimer Design 9.1 Y407M + T366I | | Descriptor: | Fc-III peptide, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
5TZJ
 
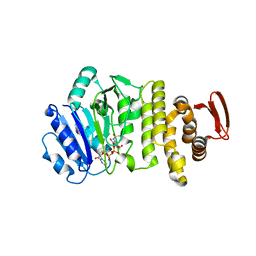 | | Crystal structure of S. aureus TarS 1-349 in complex with UDP-GlcNAc | | Descriptor: | Glycosyl transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
