3PBT
 
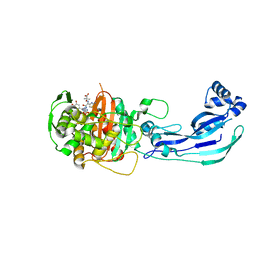 | | Crystal structure of PBP3 complexed with MC-1 | | Descriptor: | (4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-1-[({4-[(2R)-2,3-dihydroxypropyl]-3-(4,5-dihydroxypyridin-2-yl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-4-formyl-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oate, Penicillin-binding protein 3 | | Authors: | Han, S, Evdokimov, A. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBN
 
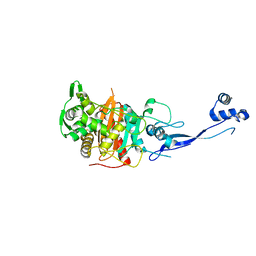 | | Crystal Structure of Apo PBP3 from Pseudomonas aeruginosa | | Descriptor: | Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBS
 
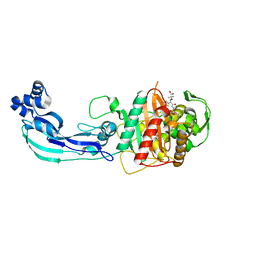 | | Crystal structure of PBP3 complexed with aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBQ
 
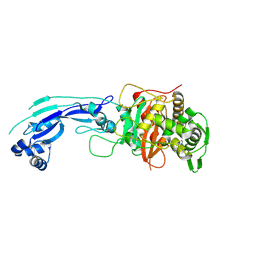 | | Crystal structure of PBP3 complexed with imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PBR
 
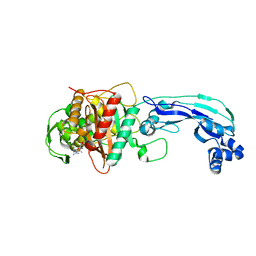 | | Crystal structure of PBP3 complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4OOM
 
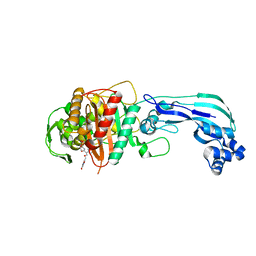 | | Crystal structure of PBP3 in complex with BAL30072 ((2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}-N-{(2S)-1-hydroxy-3-methyl-3-[(sulfooxy)amino]butan-2-yl}ethanamide) | | Descriptor: | (2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}-N-{(2S)-1-hydroxy-3-methyl-3-[(sulfooxy)amino]butan-2-yl}ethanamide, Cell division protein FtsI [Peptidoglycan synthetase] | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
4PK0
 
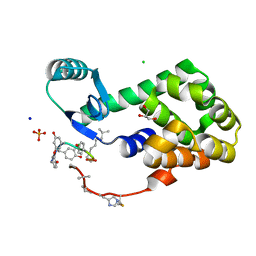 | | CRYSTAL STRUCTURE OF T4 LYSOZYME-PEPTIDE IN COMPLEX WITH TEICOPLANIN-A2-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, 8-METHYLNONANOIC ACID, ... | | Authors: | Han, S, Le, B.V, Hajare, H, Baxter, R.H.G, Miller, S.J. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure of Teicoplanin A2-2 Bound to a Catalytic Peptide Sequence via the Carrier Protein Strategy.
J.Org.Chem., 79, 2014
|
|
4PY1
 
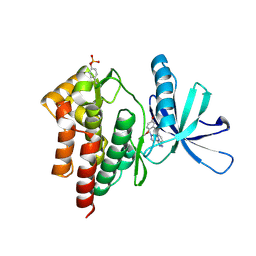 | | Crystal structure of Tyk2 in complex with compound 15, 6-((2,5-dimethoxyphenyl)thio)-3-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine | | Descriptor: | 6-[(2,5-dimethoxyphenyl)sulfanyl]-3-(1-methyl-1H-pyrazol-4-yl)[1,2,4]triazolo[4,3-b]pyridazine, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Han, S, Knafels, J.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Kinase domain inhibition of leucine rich repeat kinase 2 (LRRK2) using a [1,2,4]triazolo[4,3-b]pyridazine scaffold.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4PJZ
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME-GSS-PEPTIDE IN COMPLEX WITH TEICOPLANIN-A2-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose, 8-METHYLNONANOIC ACID, ... | | Authors: | Han, S, Le, B.V, Hajare, H, Baxter, R.H.G, Miller, S.J. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray Crystal Structure of Teicoplanin A2-2 Bound to a Catalytic Peptide Sequence via the Carrier Protein Strategy.
J.Org.Chem., 79, 2014
|
|
7WBN
 
 | | PDB structure of RevCC | | Descriptor: | RevCC | | Authors: | Han, S, Kim, D, Kaur, M, Lim, Y.B, Barnwal, R.P. | | Deposit date: | 2021-12-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pseudo-Isolated alpha-Helix Platform for the Recognition of Deep and Narrow Targets.
J.Am.Chem.Soc., 144, 2022
|
|
1T0E
 
 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*GP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*UP*CP*GP*G)-3', SULFATE ION | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1T0D
 
 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*(MTU)P*AP*GP*UP*CP*GP*CP*UP*GP*G)-3' | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
7UJ3
 
 | |
2QLU
 
 | |
4U1E
 
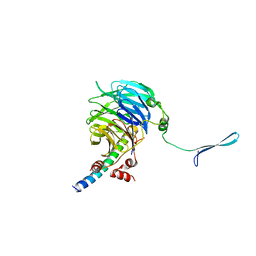 | | Crystal structure of the eIF3b-CTD/eIF3i/eIF3g-NTD translation initiation complex | | Descriptor: | Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit G, Eukaryotic translation initiation factor 3 subunit I | | Authors: | Zhang, S, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-07-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell, 158, 2014
|
|
1KNF
 
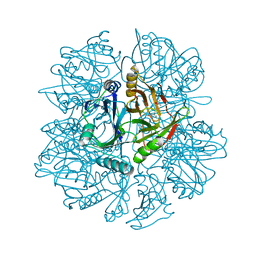 | | Crystal Structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase Complexed with 3-methyl Catechol under Anaerobic Condition | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, 3-METHYLCATECHOL, FE (II) ION, ... | | Authors: | Han, S, Bolin, J.T. | | Deposit date: | 2001-12-18 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol.
J.Biol.Chem., 273, 1998
|
|
4H1J
 
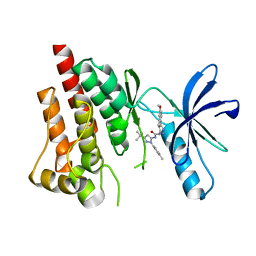 | | Crystal structure of PYK2 with the pyrazole 13a | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[3-(4-methoxy-2-methylphenyl)-1H-pyrazol-5-yl]urea, Protein-tyrosine kinase 2-beta | | Authors: | Han, S. | | Deposit date: | 2012-09-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of novel series of pyrazole and indole-urea based DFG-out PYK2 inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4HCU
 
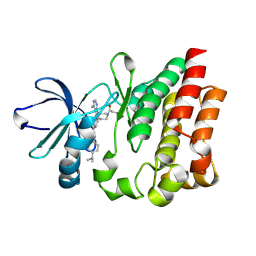 | | Crystal structure of ITK in complext with compound 40 | | Descriptor: | 3-{4-amino-1-[(3R)-1-propanoylpiperidin-3-yl]-1H-pyrazolo[3,4-d]pyrimidin-3-yl}-N-[4-(propan-2-yl)phenyl]benzamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Han, S, Caspers, N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Covalent inhibitors of interleukin-2 inducible T cell kinase (itk) with nanomolar potency in a whole-blood assay.
J.Med.Chem., 55, 2012
|
|
4FSF
 
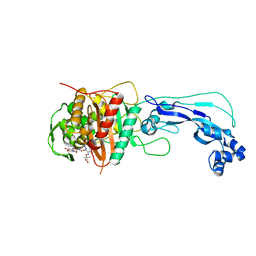 | | Crystal structure of Pseudomonas aeruginosa PBP3 complexed with compound 14 | | Descriptor: | (4R,5S,8Z)-8-(2-amino-1,3-thiazol-4-yl)-1-[3-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-1,2-oxazol-5-yl]-5-formyl-11,11-dimethyl-1,7-dioxo-4-(sulfoamino)-10-oxa-2,6,9-triazadodec-8-en-12-oic acid, Penicillin-binding protein 3 | | Authors: | Han, S. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel monobactams utilizing a siderophore uptake mechanism for the treatment of gram-negative infections.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1KND
 
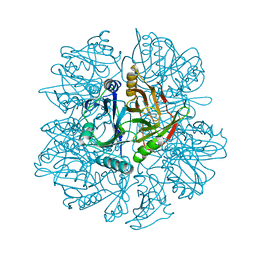 | | Crystal Structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase Complexed with Catechol under Anaerobic Condition | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, CATECHOL, FE (II) ION, ... | | Authors: | Han, S, Bolin, J.T. | | Deposit date: | 2001-12-18 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol.
J.Biol.Chem., 273, 1998
|
|
4H1M
 
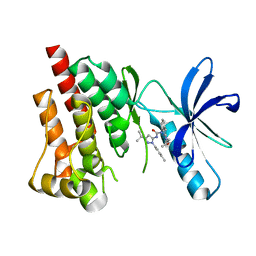 | | Crystal structure of PYK2 with the indole 10c | | Descriptor: | 7-({[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]carbamoyl}amino)-N-(propan-2-yl)-1H-indole-2-carboxamide, Protein-tyrosine kinase 2-beta | | Authors: | Han, S. | | Deposit date: | 2012-09-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Identification of novel series of pyrazole and indole-urea based DFG-out PYK2 inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4IO9
 
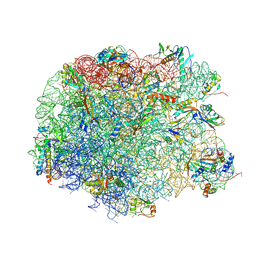 | | Crystal structure of compound 4d bound to large ribosomal subunit (50S) from Deinococcus radiodurans | | Descriptor: | (3aS,4R,7R,8S,9S,10R,11R,13R,15R,15aR)-4-ethyl-11-methoxy-3a,7,9,11,13,15-hexamethyl-2,6,14-trioxo-10-{[3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranosyl]oxy}tetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-8-yl (2R)-2-(pyridin-3-yl)pyrrolidine-1-carboxylate, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | Authors: | Han, S, Marr, E.S. | | Deposit date: | 2013-01-07 | | Release date: | 2013-03-06 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Novel 3-O-carbamoyl erythromycin A derivatives (carbamolides) with activity against resistant staphylococcal and streptococcal isolates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4IOA
 
 | | Crystal structure of compound 4e bound to large ribosomal subunit (50S) from Deinococcus radiodurans | | Descriptor: | (3aS,4R,7R,8S,9S,10R,11R,13R,15R,15aR)-4-ethyl-11-methoxy-3a,7,9,11,13,15-hexamethyl-2,6,14-trioxo-10-{[3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranosyl]oxy}tetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-8-yl (2R)-2-[4-(acetylamino)phenyl]-2,3-dihydro-1H-pyrrole-1-carboxylate, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | Authors: | Han, S, Marr, E.S. | | Deposit date: | 2013-01-07 | | Release date: | 2013-03-06 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Novel 3-O-carbamoyl erythromycin A derivatives (carbamolides) with activity against resistant staphylococcal and streptococcal isolates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4IOC
 
 | | Crystal structure of compound 4f bound to large ribosomal subunit (50S) from Deinococcus radiodurans | | Descriptor: | (3aS,4R,7R,8S,9S,10R,11R,13R,15S,15aR)-4-ethyl-11-methoxy-3a,7,9,11,13,15-hexamethyl-2,6,14-trioxo-10-{[3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranosyl]oxy}tetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-8-yl (2R)-4,4-dimethyl-2-(pyridin-3-yl)pyrrolidine-1-carboxylate, 23S ribosomal RNA, 50S ribosomal protein L11, ... | | Authors: | Han, S, Marr, E.S. | | Deposit date: | 2013-01-07 | | Release date: | 2013-03-06 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Novel 3-O-carbamoyl erythromycin A derivatives (carbamolides) with activity against resistant staphylococcal and streptococcal isolates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1KMY
 
 | | Crystal Structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase Complexed with 2,3-dihydroxybiphenyl under Anaerobic Condition | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, BIPHENYL-2,3-DIOL, FE (II) ION, ... | | Authors: | Han, S, Bolin, J.T. | | Deposit date: | 2001-12-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol.
J.Biol.Chem., 273, 1998
|
|
