7XL6
 
 | |
7UZM
 
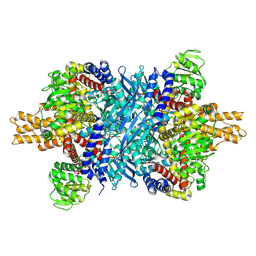 | | Glutamate dehydrogenase 1 from human liver | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Zhang, Z. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
6J9R
 
 | |
7VGR
 
 | |
7VGS
 
 | |
7XME
 
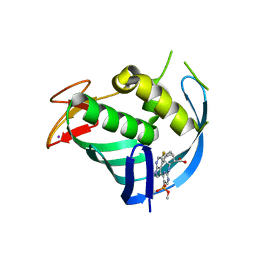 | | Structure of Influenza A virus polymerase basic protein 2 (PB2) with an azazindole derivative | | Descriptor: | (2~{S},3~{S})-3-[[5-dimethoxyphosphoryl-4-(5-fluoranyl-1~{H}-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-2-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, IODIDE ION, Polymerase basic protein 2 | | Authors: | Zhang, Z. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Discovery of a novel azaindole derivatives targeting the influenza PB2 cap binding region
To Be Published
|
|
5DAB
 
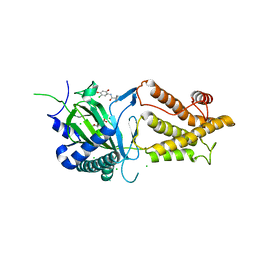 | | Crystal structure of FTO-IN115 | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase FTO, CHLORIDE ION, ... | | Authors: | Chai, J, Zhou, B, Liu, W, Han, Z, Niu, T. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of FTO-IN115
To Be Published
|
|
2G9A
 
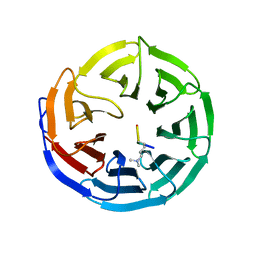 | |
2G99
 
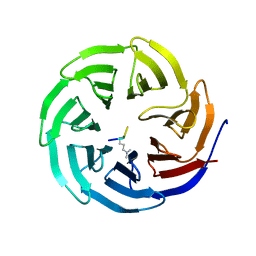 | |
5VF3
 
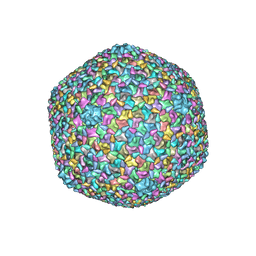 | | Bacteriophage T4 isometric capsid | | Descriptor: | Capsid vertex protein gp24, Highly immunogenic outer capsid protein, Major capsid protein, ... | | Authors: | Chen, Z, Sun, L, Zhang, Z, Fokine, A, Padilla-Sanchez, V, Hanein, D, Jiang, W, Rossmann, M.G, Rao, V.B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 isometric head at 3.3- angstrom resolution and its relevance to the assembly of icosahedral viruses.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8S4G
 
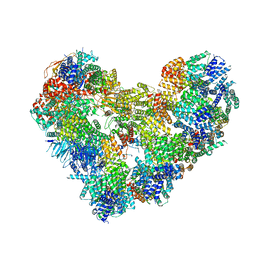 | | Cryo-EM structure of the Anaphase-promoting complex/cyclosome (APC/C) bound to co-activator Cdh1 at 3.2 Angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2024-02-21 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | New structural features of the APC/C revealed by high resolution cryo-EM structures of apo-APC/C and the APC/C-CDH1-EMI1 complex
To Be Published
|
|
6QLD
 
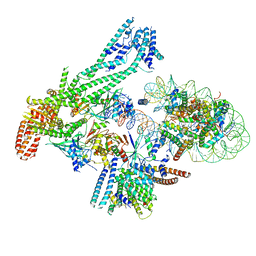 | | Structure of inner kinetochore CCAN-Cenp-A complex | | Descriptor: | DNA (125-MER), Histone H2A.1, Histone H2B.1, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
6QLF
 
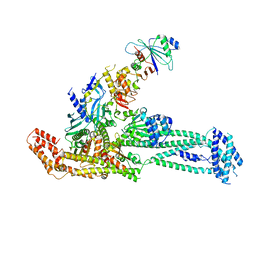 | | Structure of inner kinetochore CCAN complex with mask1 | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CHL4, Inner kinetochore subunit CTF19, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
5VRF
 
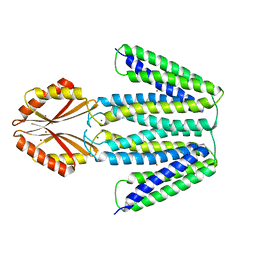 | | CryoEM Structure of the Zinc Transporter YiiP from helical crystals | | Descriptor: | Cadmium and zinc efflux pump FieF, ZINC ION | | Authors: | Coudray, N, Lopez-Redondo, M, Zhang, Z, Alexopoulos, J, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | Deposit date: | 2017-05-10 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for the alternating access mechanism of the cation diffusion facilitator YiiP.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7Y6I
 
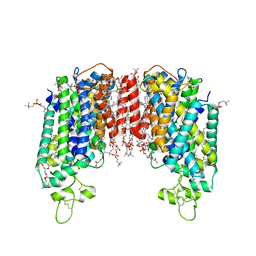 | | Cryo-EM structure of human sodium-chloride cotransporter | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nan, J, Yang, X, Shan, Z, Yuan, Y, Zhang, Y.Q. | | Deposit date: | 2022-06-20 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the human sodium-chloride cotransporter NCC.
Sci Adv, 8, 2022
|
|
7XR4
 
 | |
7XR6
 
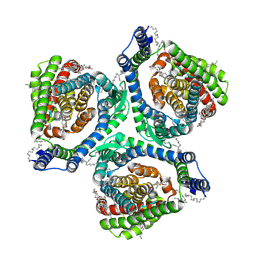 | | Structure of human excitatory amino acid transporter 2 (EAAT2) in complex with WAY-213613 | | Descriptor: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Zhao, Y, Zhang, Z. | | Deposit date: | 2022-05-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of ligand binding modes of human EAAT2.
Nat Commun, 13, 2022
|
|
4FQ7
 
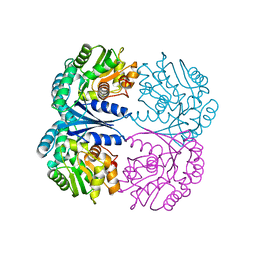 | | Crystal structure of the maleate isomerase Iso from Pseudomonas putida S16 | | Descriptor: | Maleate cis-trans isomerase | | Authors: | Lu, Y, Chen, D, Zhang, Z, Li, Q, Wu, G, Xu, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside.
Mol.Microbiol., 87, 2013
|
|
4FQ5
 
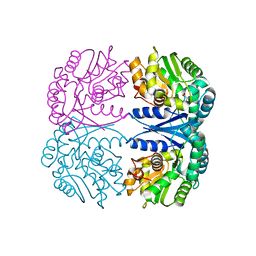 | | Crystal structure of the maleate isomerase Iso(C200A) from Pseudomonas putida S16 with maleate | | Descriptor: | MALEIC ACID, Maleate cis-trans isomerase | | Authors: | Lu, Y, Chen, D, Zhang, Z, Li, Q, Wu, G, Xu, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside.
Mol.Microbiol., 87, 2013
|
|
7Y1T
 
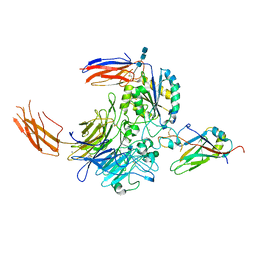 | | Complex of integrin alphaV/beta8 and L-TGF-beta1 at a ratio of 1:2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duan, Z, Zhang, Z. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Specificity of TGF-beta 1 signal designated by LRRC33 and integrin alpha V beta 8.
Nat Commun, 13, 2022
|
|
7Y1R
 
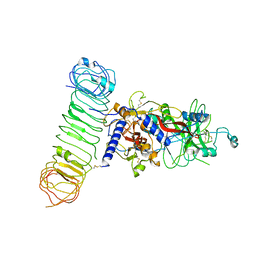 | | Human L-TGF-beta1 in complex with the anchor protein LRRC33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duan, Z, Zhang, Z. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Specificity of TGF-beta 1 signal designated by LRRC33 and integrin alpha V beta 8.
Nat Commun, 13, 2022
|
|
5XJX
 
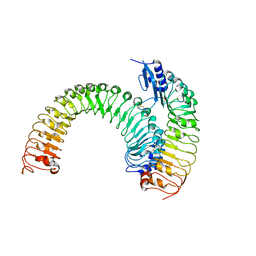 | | Pre-formed plant receptor ERL1-TMM complex | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G, Zhang, L, Han, Z, Yang, X, Liu, W, Qi, Y, Chang, J, Li, E. | | Deposit date: | 2017-05-04 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.055 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
3HH0
 
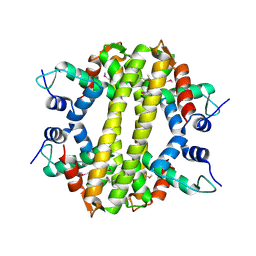 | | Crystal structure of a transcriptional regulator, MerR family from Bacillus cereus | | Descriptor: | Transcriptional regulator, MerR family | | Authors: | Palani, K, Zhang, Z, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-14 | | Release date: | 2009-05-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of a transcriptional regulator, MerR family from Bacillus cereus
To be Published
|
|
5GYY
 
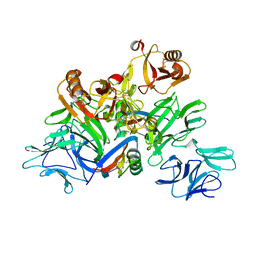 | | Plant receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S-locus protein 11, S-receptor kinase SRK9 | | Authors: | Ma, R, Han, Z, Hu, Z, Lin, G, Gong, X, Zhang, H, June, N, Chai, J. | | Deposit date: | 2016-09-24 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Plant receptor complex at 2.35 Angstroms resolution
To Be Published
|
|
5EJO
 
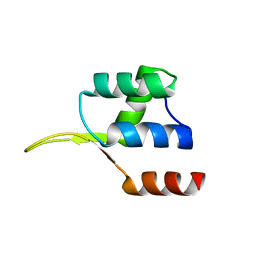 | | Crystal structure of the winged helix domain in Chromatin assembly factor 1 subunit p90 | | Descriptor: | Chromatin assembly factor 1 subunit p90 | | Authors: | Zhang, K, Gao, Y, Li, J, Burgess, R, Han, J, Liang, H, Zhang, Z, Liu, Y. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks
Nucleic Acids Res., 44, 2016
|
|
