7DHS
 
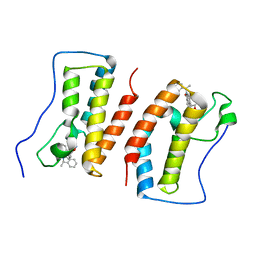 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1R)-1-phenylethyl]benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Wu, T, Xiang, Q, Wang, C, Wu, C, Zhang, C, Zhang, M, Liu, Z, Zhang, Y, Xiao, L, Xu, Y. | | Deposit date: | 2020-11-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Y06014 is a selective BET inhibitor for the treatment of prostate cancer.
Acta Pharmacol.Sin., 42, 2021
|
|
7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
7KOL
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496
to be published
|
|
4N2F
 
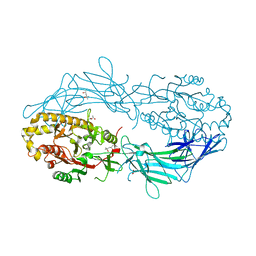 | | Crystal structure of Protein Arginine Deiminase 2 (D169A, 0 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N24
 
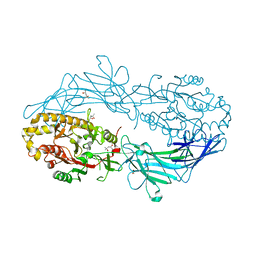 | | Crystal structure of Protein Arginine Deiminase 2 (100 uM Ca2+) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N2M
 
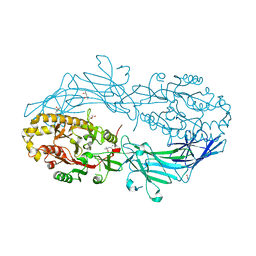 | | Crystal structure of Protein Arginine Deiminase 2 (E354A, 0 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-05 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N6F
 
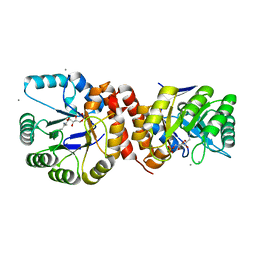 | | Crystal structure of Amycolatopsis orientalis BexX complexed with G6P | | Descriptor: | CALCIUM ION, FRUCTOSE -6-PHOSPHATE, Putative thiosugar synthase | | Authors: | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
4N2G
 
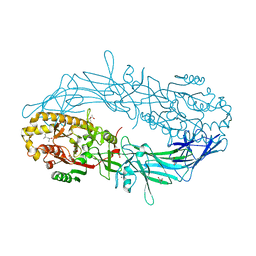 | | Crystal structure of Protein Arginine Deiminase 2 (D169A, 10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N28
 
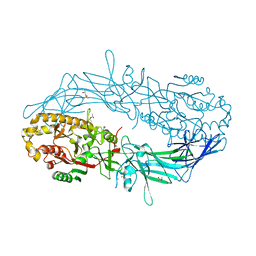 | | Crystal structure of Protein Arginine Deiminase 2 (1 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
7WDM
 
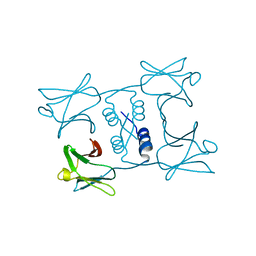 | | Fungal immunomodulatory protein FIP-gmi | | Descriptor: | Immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.123 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties
To Be Published
|
|
5XYM
 
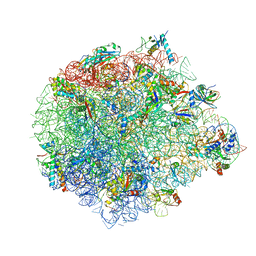 | | Large subunit of Mycobacterium smegmatis | | Descriptor: | 23S RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Li, Z, Ge, X, Zhang, Y, Zheng, L, Sanyal, S, Gao, N. | | Deposit date: | 2017-07-09 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-EM structure of Mycobacterium smegmatis ribosome reveals two unidentified ribosomal proteins close to the functional centers.
Protein Cell, 9, 2018
|
|
7XNG
 
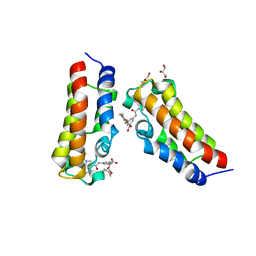 | | Crystal structure of CBP bromodomain liganded with Y08092(31g) | | Descriptor: | 3-[(1-ethanoylindol-3-yl)carbonylamino]-5-[[(2S)-oxan-2-yl]oxymethyl]benzoic acid, CREB-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08092(31g)
To Be Published
|
|
7XM7
 
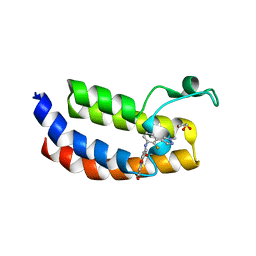 | | Crystal Structure of the CBP in complex with the Y08188 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethanoyl-~{N}-[2-fluoranyl-3-(1-methylpyrazol-4-yl)phenyl]-7-methoxy-indolizine-1-carboxamide, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as potent and selective CBP bromodomain inhibitors
To Be Published
|
|
2WG1
 
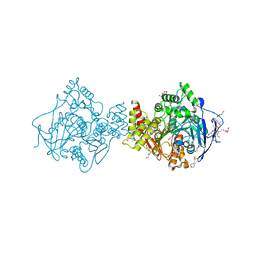 | | TERNARY COMPLEX OF THE AGED CONJUGATE OF TORPEDO CALIFORNICA ACEYLCHOLINESTERASE WITH SOMAN AND 2-PAM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Sanson, B, Nachon, F, Colletier, J.P, Froment, M.T, Toker, L, Greenblatt, H.M, Sussman, J.L, Ashani, Y, Masson, P, Silman, I, Weik, M. | | Deposit date: | 2009-04-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Snapshots of Nonaged and Aged Conjugates of Soman with Acetylcholinesterase, and of a Ternary Complex of the Aged Conjugate with Pralidoxime.
J.Med.Chem., 52, 2009
|
|
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | Descriptor: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
4K7F
 
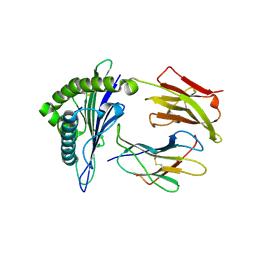 | | Newly identified epitope V60 from HBV core protein complexed with HLA-A*0201 | | Descriptor: | Beta-2-microglobulin, Core protein, HLA class I histocompatibility antigen, ... | | Authors: | Meng, S.D, Zhang, Y, Wu, Y, Qi, J.X. | | Deposit date: | 2013-04-17 | | Release date: | 2013-06-05 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The L60V variation in hepatitis B virus core protein elicits new epitope-specific cytotoxic T lymphocytes and enhances viral replication.
J.Virol., 87, 2013
|
|
7EIZ
 
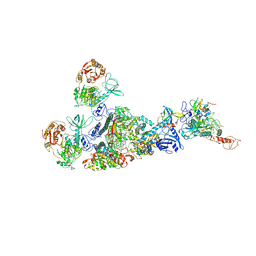 | | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Yan, L, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-22 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading
Cell, 184, 2021
|
|
4MEX
 
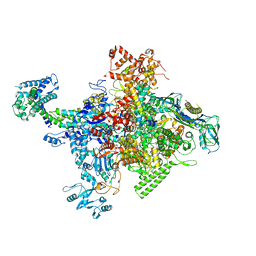 | | Crystal structure of Escherichia coli RNA polymerase in complex with salinamide A | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Feng, Y, Zhang, Y, Arnold, E, Ebright, R.H. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (3.902 Å) | | Cite: | Transcription inhibition by the depsipeptide antibiotic salinamide A.
Elife, 3, 2014
|
|
4I48
 
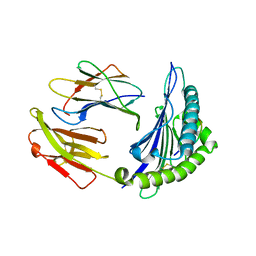 | | Structure of HLA-A68 complexed with an HIV Env derived peptide | | Descriptor: | 9-mer peptide from Envelope glycoprotein gp160, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-27 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
4IMI
 
 | | Novel Modifications on C-terminal Domain of RNA Polymerase II can Fine- tune the Phosphatase Activity of Ssu72. | | Descriptor: | CG14216, CTD, PHOSPHATE ION, ... | | Authors: | Luo, Y, Yogesha, S.D, Zhang, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-08-07 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel Modifications on C-terminal Domain of RNA Polymerase II Can Fine-tune the Phosphatase Activity of Ssu72.
Acs Chem.Biol., 8, 2013
|
|
7EU0
 
 | | The cryo-EM structure of A. thaliana Pol IV-RDR2 backtracked complex | | Descriptor: | DNA (33-MER), DNA (5'-D(*CP*TP*GP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*GP*AP*GP*TP*G)-3'), DNA-directed RNA polymerase IV subunit 1, ... | | Authors: | Fang, C.L, Wu, X.X, Huang, K, Zhang, Y. | | Deposit date: | 2021-05-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Pol IV and RDR2: A two-RNA-polymerase machine that produces double-stranded RNA.
Science, 374, 2021
|
|
3TCZ
 
 | | Human Pin1 bound to cis peptidomimetic inhibitor | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, N~2~-({(1R,2Z)-2-[(2R)-2-(formylamino)-3-(phosphonooxy)propylidene]cyclopentyl}carbonyl)-L-argininamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Kinetic Analysis of Prolyl-isomerization/Phosphorylation Cross-Talk in the CTD Code.
Acs Chem.Biol., 7, 2012
|
|
7CAL
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Inwardly Rectifying Potassium Channel KAT1 from Arabidopsis | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Potassium channel KAT1 | | Authors: | Li, S.Y, Yang, F, Sun, D.M, Zhang, Y, Zhang, M.G, Zhou, P, Liu, S.L, Zhang, Y.N, Zhang, L.H, Tian, C.L. | | Deposit date: | 2020-06-09 | | Release date: | 2020-07-29 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis.
Cell Res., 30, 2020
|
|
7CKQ
 
 | |
4OHR
 
 | | Crystal structure of MilB from Streptomyces rimofaciens | | Descriptor: | CMP/hydroxymethyl CMP hydrolase | | Authors: | Zhao, G, Zhang, Y, Liu, G, Wu, G, He, X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the N-glycosidase MilB in complex with hydroxymethyl CMP reveals its Arg23 specifically recognizes the substrate and controls its entry
Nucleic Acids Res., 42, 2014
|
|
