7XQE
 
 | | Crystal Structure of human RORgamma (C455E) LBD in complex with compound XY039 | | Descriptor: | 2,4-difluoro-N-(1-((4-(trifluoromethyl)benzyl)sulfonyl)-1,2,3,4-tetrahydroquinolin-7-yl)benzenesulfonamide, ETHANOL, GLYCEROL, ... | | Authors: | Wu, X, Li, C, Zhang, Y, Xu, Y. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery and pharmacological characterization of 1,2,3,4-tetrahydroquinoline derivatives as ROR gamma inverse agonists against prostate cancer.
Acta Pharmacol.Sin., 2024
|
|
7VCF
 
 | | Cryo-EM structure of Chlamydomonas TOC-TIC supercomplex | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, J, Yan, Z, Jin, Z, Zhang, Y. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of a TOC-TIC supercomplex spanning two chloroplast envelope membranes.
Cell, 185, 2022
|
|
7VYU
 
 | |
7XNG
 
 | | Crystal structure of CBP bromodomain liganded with Y08092(31g) | | Descriptor: | 3-[(1-ethanoylindol-3-yl)carbonylamino]-5-[[(2S)-oxan-2-yl]oxymethyl]benzoic acid, CREB-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08092(31g)
To Be Published
|
|
7XM7
 
 | | Crystal Structure of the CBP in complex with the Y08188 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethanoyl-~{N}-[2-fluoranyl-3-(1-methylpyrazol-4-yl)phenyl]-7-methoxy-indolizine-1-carboxamide, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as potent and selective CBP bromodomain inhibitors
To Be Published
|
|
7VMY
 
 | | Crystal structure of LimF prenyltransferase bound with GSPP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, ... | | Authors: | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
7VMW
 
 | | Crystal structure of LimF prenyltransferase bound with a peptide substrate and GSPP | | Descriptor: | GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | Authors: | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
7VLA
 
 | | Cryo-EM structure of the CCL15(27-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(27-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL8
 
 | | Cryo-EM structure of the Apo CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL9
 
 | | Cryo-EM structure of the CCL15(26-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(26-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7Y35
 
 | | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex | | Descriptor: | Abaloparatide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhai, X, Mao, C, Shen, Q, Zang, S, Shen, D, Zhang, H, Chen, Z, Wang, G, Zhang, C, Zhang, Y, Liu, Z. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex
To Be Published
|
|
7Y36
 
 | | Cryo-EM structure of the Teriparatide-bound human PTH1R-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, HYDROXIDE ION, ... | | Authors: | Zhai, X, Mao, C, Shen, Q, Zang, S, Shen, D, Zhang, H, Chen, Z, Wang, G, Zhang, C, Zhang, Y, Liu, Z. | | Deposit date: | 2022-06-10 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the Teriparatide-bound human PTH1R-Gs complex
to be published
|
|
7XTQ
 
 | | Cryo-EM structure of the R399-bound GPBAR-Gs complex | | Descriptor: | G-protein coupled bile acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, L, Yang, F, Wu, X, Mao, C, Sun, J, Yu, X, Zhang, Y, Zhang, P. | | Deposit date: | 2022-05-17 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis and molecular mechanism of biased GPBAR signaling in regulating NSCLC cell growth via YAP activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1GN4
 
 | | H145E mutant of Mycobacterium tuberculosis iron-superoxide dismutase. | | Descriptor: | MANGANESE (II) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Badasso, M.O, Tickle, I.J, Newton, M, Wood, S.P, Zhang, Y, Young, D.B. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineering a Change in the Metal-Ion Specificity of the Iron-Depedent Superoxide Dismutase from Mycobacterium Tuberculosis. X-Ray Structure Analysis of Site-Directed Mutants
Eur.J.Biochem., 251, 1998
|
|
1GN3
 
 | | H145Q mutant of Mycobacterium tuberculosis iron-superoxide dismutase. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE | | Authors: | Bunting, K.A, Cooper, J.B, Badasso, M.O, Tickle, I.J, Newton, M, Wood, S.P, Zhang, Y, Young, D.B. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Engineering a Change in the Metal-Ion Specificity of the Iron-Depedent Superoxide Dismutase from Mycobacterium Tuberculosis. X-Ray Structure Analysis of Site-Directed Mutants.
Eur.J.Biochem., 251, 1998
|
|
5Y1H
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,4-difluorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2R)-2-[[2,4-bis(fluoranyl)phenyl]methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,4-difluorobenzyl)ureido)-N-hydroxy-4-methylpentanamide
To Be Published
|
|
5Y1R
 
 | |
7VLB
 
 | |
5Y3I
 
 | |
5Y1S
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3,4-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2R)-2-[(3,4-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, (2S)-2-[(3,4-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3,4-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide
To Be Published
|
|
5Y1V
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,6-diethylphenyl)ureido)- N-hydroxy-4-methylpentanamide | | Descriptor: | (2R)-2-[(2,6-diethylphenyl)carbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,6-diethylphenyl)ureido)- N-hydroxy-4-methylpentanamide
To Be Published
|
|
5Y19
 
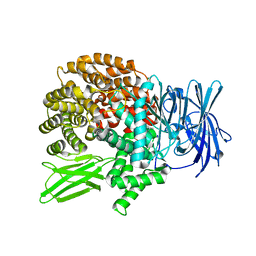 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-((2S,3R)-3-amino-2-hydroxy-4-phenylbutanamido)- N-hydroxy-4-methylpentanamide. | | Descriptor: | (2S)-2-[[(2S,3R)-3-azanyl-2-oxidanyl-4-phenyl-butanoyl]amino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-((2S,3R)-3-amino-2-hydroxy-4-phenylbutanamido)- N-hydroxy-4-methylpentanamide.
To Be Published
|
|
7YE1
 
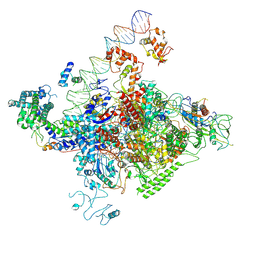 | | The cryo-EM structure of C. crescentus GcrA-TACup | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (57-MER)-non template, DNA (57-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
7YE2
 
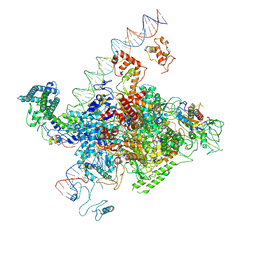 | | The cryo-EM structure of C. crescentus GcrA-TACdown | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (90-MER)-non template, DNA (90-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
4M85
 
 | |
