8XI5
 
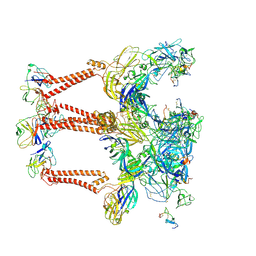 | | Structure of Eastern Equine Encephalitis VLP in complex with the receptor VLDLR LA3-5 | | Descriptor: | CALCIUM ION, Capsid protein, Spike glycoprotein E1, ... | | Authors: | Cao, D, Ma, B, Cao, Z, Xu, X, Zhang, X, Xiang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The receptor VLDLR binds Eastern Equine Encephalitis virus through multiple distinct modes.
Nat Commun, 15, 2024
|
|
8AJ8
 
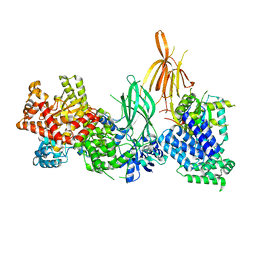 | | Structure of p110 gamma bound to the p84 regulatory subunit | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, Phosphoinositide 3-kinase regulatory subunit 6 | | Authors: | Burke, J.E, Williams, R.L, Zhang, X. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (8.5 Å) | | Cite: | Molecular basis for differential activation of p101 and p84 complexes of PI3K gamma by Ras and GPCRs.
Cell Rep, 42, 2023
|
|
8IZB
 
 | | Lysophosphatidylserine receptor GPR174-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gong, W, Liu, G, Li, X, Zhang, X. | | Deposit date: | 2023-04-06 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
4GZN
 
 | | Mouse ZFP57 zinc fingers in complex with methylated DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Liu, Y, Zhang, X, Cheng, X. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence.
Genes Dev., 26, 2012
|
|
4GZU
 
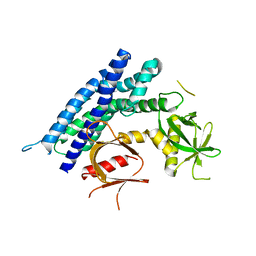 | | Crystal structure of the DH-PH-PH domain of FARP2 | | Descriptor: | FERM, RhoGEF and pleckstrin domain-containing protein 2 | | Authors: | He, X, Zhang, X. | | Deposit date: | 2012-09-06 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Autoinhibition of the Guanine Nucleotide Exchange Factor FARP2.
Structure, 21, 2013
|
|
8XKV
 
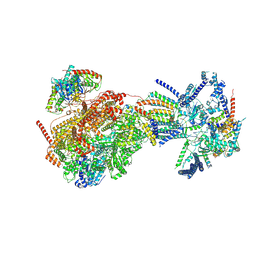 | | Cryo-EM structure of the Ycf2-FtsHi motor complex from Arabidopsis in Apo state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, ATP-dependent zinc metalloprotease FTSH 12, chloroplastic, ... | | Authors: | Liang, K, Zhan, X, Xu, Q, Wu, J, Yan, Z. | | Deposit date: | 2023-12-25 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the chloroplast protein import in land plants.
Cell, 187, 2024
|
|
4H6Y
 
 | | Crystal structure of the DH-PH-PH domain of FARP1 | | Descriptor: | FERM, RhoGEF and pleckstrin domain-containing protein 1 | | Authors: | He, X, Zhang, X. | | Deposit date: | 2012-09-19 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.09 Å) | | Cite: | Structural Basis for Autoinhibition of the Guanine Nucleotide Exchange Factor FARP2.
Structure, 21, 2013
|
|
1RHP
 
 | |
3O05
 
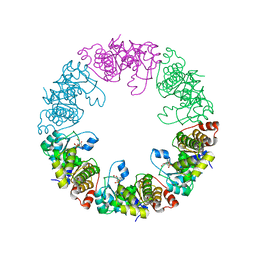 | | Crystal Structure of Yeast Pyridoxal 5-Phosphate Synthase Snz1 Complxed with Substrate PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxine biosynthesis protein SNZ1 | | Authors: | Teng, Y.B, Zhang, X, He, Y.X, Hu, H.X, Zhou, C.Z. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the catalytic mechanism of the yeast pyridoxal 5-phosphate synthase Snz1
Biochem.J., 432, 2010
|
|
4EW0
 
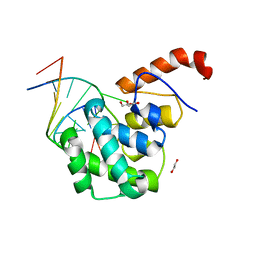 | | mouse MBD4 glycosylase domain in complex with a G:5hmU (5-hydroxymethyluracil) mismatch | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*(5HU)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
7WQR
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 28 | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ~{N}-(3-chlorophenyl)-2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]benzamide | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQM
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 24 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(2-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQS
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 25 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(3-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
5YC6
 
 | | The crystal structure of uPA in complex with 4-Bromobenzylamirne at pH4.6 | | Descriptor: | 1-(4-BROMOPHENYL)METHANAMINE, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Zhang, X, Huang, M.D. | | Deposit date: | 2017-09-06 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Halogen bonding for the design of inhibitors by targeting the S1 pocket of serine proteases
Rsc Adv, 8, 2018
|
|
7XKG
 
 | | Crystal structure of an intramolecular mesacyl-CoA transferase from the 3-hydroxypropionic acid cycle of Roseiflexus castenholzii | | Descriptor: | Acyl-CoA transferase/carnitine dehydratase-like protein | | Authors: | Min, Z.Z, Fan, C.P, Wu, W.P, Xin, Y.Y, Liu, M.H, Zhang, X, Wang, Z.G, Xu, X.L. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Intramolecular Mesaconyl-Coenzyme A Transferase From the 3-Hydroxypropionic Acid Cycle of Roseiflexus castenholzii .
Front Microbiol, 13, 2022
|
|
6IJJ
 
 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
4F0Q
 
 | | MspJI Restriction Endonuclease - P21 Form | | Descriptor: | MAGNESIUM ION, Restriction endonuclease | | Authors: | Horton, J.R, Mabuchi, M, Cohen-Karni, D, Zhang, X, Griggs, R, Samaranayake, M, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2012-05-04 | | Release date: | 2012-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
4F0P
 
 | | MspJI Restriction Endonuclease - P31 Form | | Descriptor: | MAGNESIUM ION, Restriction endonuclease | | Authors: | Horton, J.R, Mabuchi, M, Cohen-Karni, D, Zhang, X, Griggs, R, Samaranayake, M, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2012-05-04 | | Release date: | 2012-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
3JA7
 
 | | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution | | Descriptor: | Portal protein gp20 | | Authors: | Sun, L, Zhang, X, Gao, S, Rao, P.A, Padilla-Sanchez, V, Chen, Z, Sun, S, Xiang, Y, Subramaniam, S, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution.
Nat Commun, 6, 2015
|
|
8HPO
 
 | | Cryo-EM structure of a SIN3/HDAC complex from budding yeast | | Descriptor: | Histone deacetylase RPD3, PHOSPHOTHREONINE, POTASSIUM ION, ... | | Authors: | Guo, Z, Zhan, X, Wang, C. | | Deposit date: | 2022-12-12 | | Release date: | 2023-05-03 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a SIN3-HDAC complex from budding yeast.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2M5L
 
 | |
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
7D7N
 
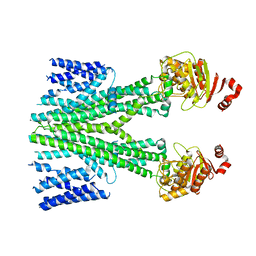 | | Cryo-EM structure of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
