5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
8GOE
 
 | | Structure of hSLC19A1+5-MTHF | | Descriptor: | N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-08-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
8GOF
 
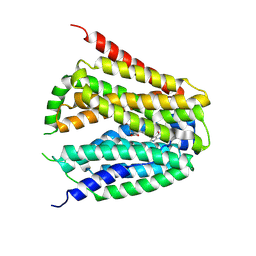 | | Structure of hSLC19A1+PMX | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-08-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
5OAF
 
 | | Human Rvb1/Rvb2 heterohexamer in INO80 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Aramayo, R.J, Bythell-Douglas, R, Ayala, R, Willhoft, O, Wigley, D, Zhang, X. | | Deposit date: | 2017-06-21 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Cryo-EM structures of the human INO80 chromatin-remodeling complex.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3AIY
 
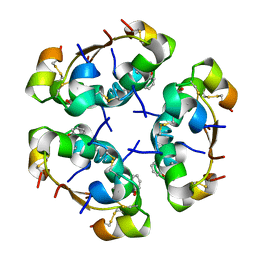 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, REFINED AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
4DEN
 
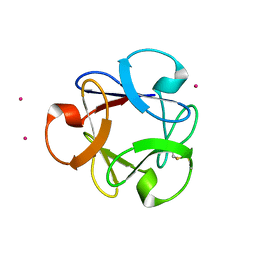 | | Structural insightsinto potent, specific anti-HIV property of actinohivin; Crystal structure of actinohivin in complex with alpha(1-2) mannobiose moiety of high-mannose type glycan of gp120 | | Descriptor: | Actinohivin, POTASSIUM ION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-01-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the specific anti-HIV property of actinohivin: structure of its complex with the alpha(1–2)mannobiose moiety of gp120
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6U9I
 
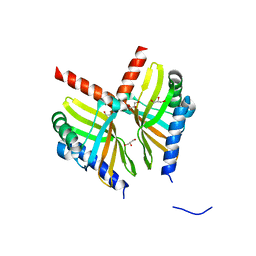 | | Crystal structure of BvnE pinacolase from Penicillium brevicompactum | | Descriptor: | BvnE, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Ye, Y, Du, L, Zhang, X, Newmister, S.A, McCauley, M, Alegre-Requena, J.V, Zhang, W, Mu, S, Minami, A, Fraley, A.E, Adrover-Castellano, M.L, Carney, N, Shende, V.V, Oikawa, H, Kato, H, Tsukamoto, S, Paton, R.S, Williams, R.M, Sherman, D.H, Li, S. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | Fungal-derived brevianamide assembly by a stereoselective semipinacolase.
Nat Catal, 3, 2020
|
|
6X19
 
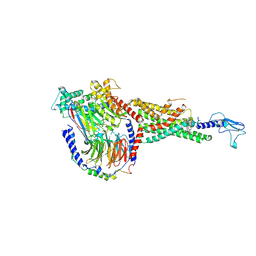 | | Non peptide agonist CHU-128, bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | 3-{(1S)-1-[5-(2,2-dimethylmorpholin-4-yl)-2-{(4S)-2-(4-fluoro-3,5-dimethylphenyl)-3-[3-(4-fluoro-1-methyl-1H-indazol-5-yl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]-4-methyl-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carbonyl}-1H-indol-1-yl]butyl}-1,2,4-oxadiazol-5(4H)-one, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
6X18
 
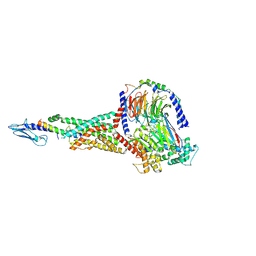 | | GLP-1 peptide hormone bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | Glucagon, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
7YG8
 
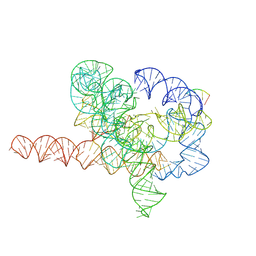 | | Cryo-EM structure of Tetrahymena ribozyme conformation 5 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (387-MER), RNA (5'-R(*CP*CP*CP*UP*C)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
8JEL
 
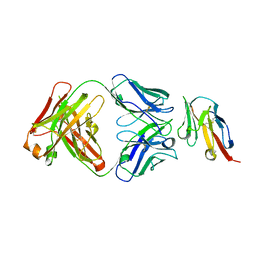 | | Crystal structure of TIGIT in complexed with Ociperlimab, crystal form I | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
7YG9
 
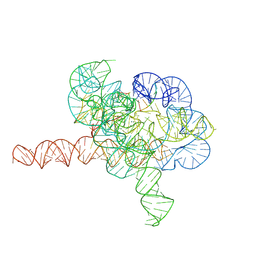 | | Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (391-MER), RNA (5'-R(*CP*CP*CP*UP*CP*U)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGA
 
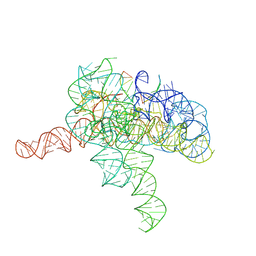 | | Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*U)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGD
 
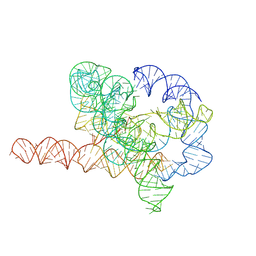 | | Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (384-MER), RNA (5'-R(*CP*C)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGC
 
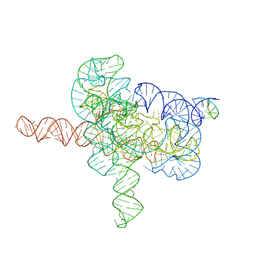 | | Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*UP*UP*AP*AP*CP*C)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
3KH8
 
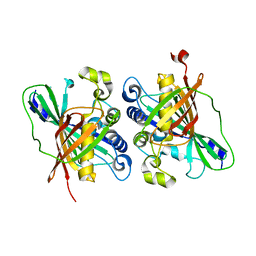 | | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici | | Descriptor: | MaoC-like dehydratase | | Authors: | Wang, H, Zhang, K, Guo, J, Zhou, Q, Zheng, X, Sun, F, Pang, H, Zhang, X. | | Deposit date: | 2009-10-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici
To be Published
|
|
8JEN
 
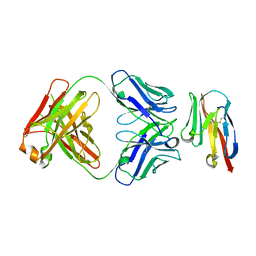 | | Crystal structure of TIGIT in complexed with Ociperlimab, crystal form II | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
6R9B
 
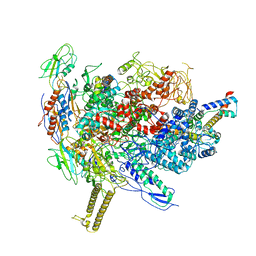 | |
4V40
 
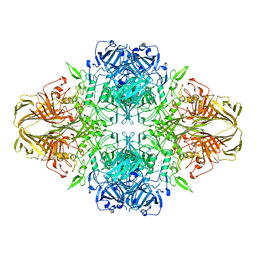 | | BETA-GALACTOSIDASE | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Jacobson, R.H, Zhang, X, Dubose, R.F, Matthews, B.W. | | Deposit date: | 1994-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of beta-galactosidase from E. coli.
Nature, 369, 1994
|
|
6LOM
 
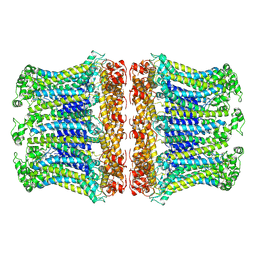 | |
4C6T
 
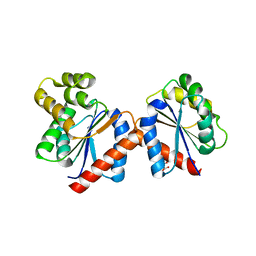 | | Crystal structure of the RPS4 and RRS1 TIR domain heterodimer | | Descriptor: | DISEASE RESISTANCE PROTEIN RPS4, MALONIC ACID, PROBABLE WRKY TRANSCRIPTION FACTOR 52 | | Authors: | Williams, S.J, Sohn, K.H, Wan, L, Bernoux, M, Ma, Y, Segonzac, C, Ve, T, Sarris, P, Ericsson, D.J, Saucet, S.B, Zhang, X, Parker, J, Dodds, P.N, Jones, J.D.G, Kobe, B. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor.
Science, 344, 2014
|
|
2QJT
 
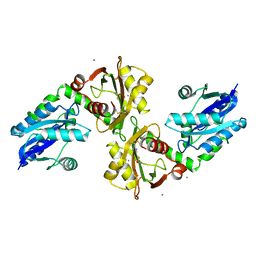 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase complexed with AMP and MN ion from Francisella tularensis | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-09 | | Release date: | 2008-03-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
6EDR
 
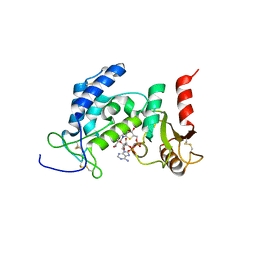 | | Crystal Structure of Human CD38 in Complex with 4'-Thioribose NAD+ | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)thiolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Dai, Z, Zhang, X.N, Nasertorabi, F, Cheng, Q, Pei, H, Louie, S.G, Stevens, C.R, Zhang, Y. | | Deposit date: | 2018-08-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Facile chemoenzymatic synthesis of a novel stable mimic of NAD.
Chem Sci, 9, 2018
|
|
8BQN
 
 | |
8YS2
 
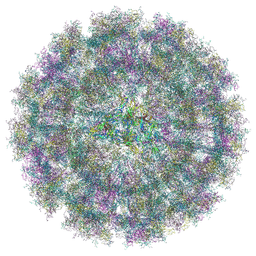 | | Overall structure of Eastern Equine Encephalitis virus VLP in complex with the receptor VLDLR LA1-2 | | Descriptor: | CALCIUM ION, Capsid protein, Spike glycoprotein E1, ... | | Authors: | Cao, D, Ma, B, Cao, Z, Xu, X, Zhang, X, Xiang, Y. | | Deposit date: | 2024-03-22 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | The receptor VLDLR binds Eastern Equine Encephalitis virus through multiple distinct modes.
Nat Commun, 15, 2024
|
|
