7QFT
 
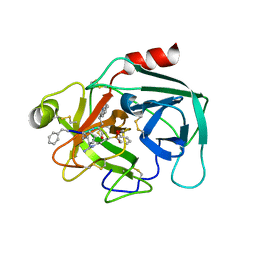 | | Crystal structure of KLK6 in complex with compound 16a | | Descriptor: | KLK6 Activity-Based Probe (Ahx-DPhe-Cha-Dht-Arg-DPP), Kallikrein-6 | | Authors: | Jagtap, P.K.A, Zhang, L, De Vita, E, Tate, E.W, Hennig, J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A KLK6 Activity-Based Probe Reveals a Role for KLK6 Activity in Pancreatic Cancer Cell Invasion.
J.Am.Chem.Soc., 144, 2022
|
|
7QFV
 
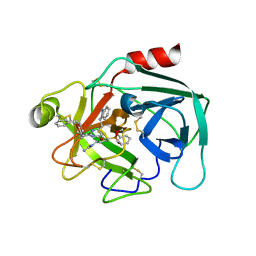 | | Crystal structure of KLK6 in complex with compound 17a | | Descriptor: | KLK6 Activity-Based Probe (Ahx-DPhe-Ser(Z)-Dht-Arg-DPP), Kallikrein-6 | | Authors: | Jagtap, P.K.A, Zhang, L, De Vita, E, Tate, E.W, Hennig, J. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A KLK6 Activity-Based Probe Reveals a Role for KLK6 Activity in Pancreatic Cancer Cell Invasion.
J.Am.Chem.Soc., 144, 2022
|
|
6E9C
 
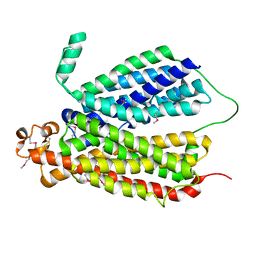 | | Selenomethionine Derivative Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
7REF
 
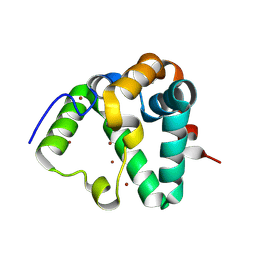 | | Structure of MS3494 from Mycobacterium smegmatis | | Descriptor: | BROMIDE ION, MS3494 | | Authors: | Kent, J.E, Aleshin, A.E, Zhang, L, Niederweis, M, Marassi, F.M. | | Deposit date: | 2021-07-12 | | Release date: | 2021-08-18 | | Last modified: | 2022-06-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A periplasmic cinched protein is required for siderophore secretion and virulence of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
9CGX
 
 | | Alzheimer's Disease Seeded 0N3R Tau Fibrils | | Descriptor: | Isoform Fetal-tau of Microtubule-associated protein tau | | Authors: | Duan, P, Dregni, A.J, Xu, H, Changolkar, L, Lee, V.M.-Y, Hong, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Alzheimer's disease seeded tau forms paired helical filaments yet lacks seeding potential.
J.Biol.Chem., 300, 2024
|
|
9CGZ
 
 | | Alzheimer's Disease Seeded Mixed 0N4R and 0N3R Tau Fibrils | | Descriptor: | Isoform Fetal-tau of Microtubule-associated protein tau | | Authors: | Duan, P, Dregni, A.J, Xu, H, Changolkar, L, Lee, V.M.-Y, Hong, M. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Alzheimer's disease seeded tau forms paired helical filaments yet lacks seeding potential.
J.Biol.Chem., 300, 2024
|
|
7SL9
 
 | | CryoEM structure of SMCT1 | | Descriptor: | Sodium-coupled monocarboxylate transporter 1, butanoic acid, nanobody Nb2 | | Authors: | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | Deposit date: | 2021-10-23 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
7SLA
 
 | | CryoEM structure of SGLT1 at 3.15 Angstrom resolution | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Sodium/glucose cotransporter 1, nanobody Nb1 | | Authors: | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | Deposit date: | 2021-10-23 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
7SL8
 
 | | CryoEM structure of SGLT1 at 3.4 A resolution | | Descriptor: | CHOLESTEROL, Sodium/glucose cotransporter 1, nanobody Nb1 | | Authors: | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | Deposit date: | 2021-10-23 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
7E6T
 
 | | Structural insights into the activation of human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYCLOMETHYLTRYPTOPHAN, ... | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
7E6U
 
 | | the complex of inactive CaSR and NB2D11 | | Descriptor: | Extracellular calcium-sensing receptor, NB-2D11 | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
5B5K
 
 | | Crystal structure of Izumo1, the mammalian sperm ligand for egg Juno | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Izumo sperm-egg fusion protein 1 | | Authors: | Nishimura, K, Han, L, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-05-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of sperm Izumo1 reveals unexpected similarities with Plasmodium invasion proteins.
Curr.Biol., 26, 2016
|
|
6M54
 
 | | Human apo ferritin frozen on TEM grid with Amorphous nickel titanium alloy supporting film | | Descriptor: | FE (II) ION, Ferritin heavy chain | | Authors: | Huang, X, Zhang, L, Wen, Z, Chen, H, Li, S, Ji, G, Yin, C, Sun, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation.
Prog.Biophys.Mol.Biol., 156, 2020
|
|
3KBJ
 
 | |
4MKI
 
 | | Cobalt transporter ATP-binding subunit | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Energy-coupling factor transporter ATP-binding protein EcfA2, SULFATE ION | | Authors: | Yu, Y, Zhang, L, Chai, C.L, Heymann, D. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a homodimeric ATPase subunit of an ECF transporter
Protein Cell, 4, 2013
|
|
7CUW
 
 | | Ubiquinol Binding Site of Cytochrome bo3 from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-25 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUQ
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUB
 
 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | Deposit date: | 2020-08-22 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8YJT
 
 | | Cryo-EM structure of the flagellar C ring in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-03-02 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structure of the flagellar C ring in the CCW state
To Be Published
|
|
5U8K
 
 | | RitR mutant - C128S | | Descriptor: | Response regulator | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2016-12-14 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | RitR is an archetype for a novel family of redox sensors in the streptococci that has evolved from two-component response regulators and is required for pneumococcal colonization.
PLoS Pathog., 14, 2018
|
|
8I0A
 
 | |
5AYW
 
 | | Structure of a membrane complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Huang, Y, Han, L, Zheng, J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.555 Å) | | Cite: | Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins
Nat.Struct.Mol.Biol., 23, 2016
|
|
7XA9
 
 | | Structure of Arabidopsis thaliana CLCa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Ji, S, Jin, H, Kaiming, Z, Mingxing, W, Shanshan, L, Long, C. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the plant nitrate transporter AtCLCa reveals characteristics of the anion-binding site and the ATP-binding pocket.
J.Biol.Chem., 299, 2023
|
|
5EJN
 
 | |
5II6
 
 | | Crystal structure of the ZP-N1 domain of mouse sperm receptor ZP2 at 0.95 A resolution | | Descriptor: | Zona pellucida sperm-binding protein 2 | | Authors: | Dioguardi, E, Han, L, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
