8Z5C
 
 | |
8Z5D
 
 | |
8Z5E
 
 | |
9C49
 
 | |
8Z5F
 
 | |
6TGC
 
 | | CryoEM structure of the ternary DOCK2-ELMO1-RAC1 complex. | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
7N61
 
 | | structure of C2 projections and MIPs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAP147, FAP178, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7N6G
 
 | | C1 of central pair | | Descriptor: | CPC1, Calmodulin, DPY30, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-08 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1HQ3
 
 | | CRYSTAL STRUCTURE OF THE HISTONE-CORE-OCTAMER IN KCL/PHOSPHATE | | Descriptor: | CHLORIDE ION, HISTONE H2A-IV, HISTONE H2B, ... | | Authors: | Chantalat, L, Nicholson, J.M, Lambert, S.J, Reid, A.J, Donovan, M.J, Reynolds, C.D, Wood, C.M, Baldwin, J.P. | | Deposit date: | 2000-12-14 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the histone-core octamer in KCl/phosphate crystals at 2.15 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2PQE
 
 | | Solution structure of proline-free mutant of staphylococcal nuclease | | Descriptor: | Thermonuclease | | Authors: | Shan, L, Tong, Y, Xie, T, Wang, M, Wang, J. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Restricted backbone conformational and motional flexibilities of loops containing peptidyl-proline bonds dominate the enzyme activity of staphylococcal nuclease.
Biochemistry, 46, 2007
|
|
6KXW
 
 | | Crystal structure of human aquaporin AQP7 in bound to glycerol | | Descriptor: | Aquaporin-7, GLYCEROL | | Authors: | Zhang, L, Yao, D, Zhou, F, Zhang, Q, Zhou, L, Cao, Y. | | Deposit date: | 2019-09-13 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The structural basis for glycerol permeation by human AQP7
Sci Bull (Beijing), 66, 2020
|
|
5XPD
 
 | |
7XMK
 
 | | Crystal structure of human RIPK1 kinase domain in complex with compound SKLB923 | | Descriptor: | 5-[2-(cyclopropylcarbonylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-(3-fluorophenyl)ethyl]-1-methyl-indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Zhang, L, Wang, Y, Li, Y, Yang, S. | | Deposit date: | 2022-04-26 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.376 Å) | | Cite: | From Hit to Lead: Structure-Based Optimization of Novel Selective Inhibitors of Receptor-Interacting Protein Kinase 1 (RIPK1) for the Treatment of Inflammatory Diseases.
J.Med.Chem., 67, 2024
|
|
6LKQ
 
 | | The Structural Basis for Inhibition of Ribosomal Translocation by Viomycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, L, Wang, Y.H, Lancaster, L, Zhou, J, Noller, H.F. | | Deposit date: | 2019-12-20 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for inhibition of ribosomal translocation by viomycin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8JU8
 
 | | de novo designed protein | | Descriptor: | de novo designed protein | | Authors: | Zhang, L. | | Deposit date: | 2023-06-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | de novo designed Rossmann fold protein
To Be Published
|
|
7N3O
 
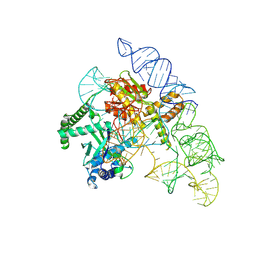 | | Cryo-EM structure of the Cas12k-sgRNA complex | | Descriptor: | Cas12k, Single guide RNA | | Authors: | Chang, L, Li, Z, Xiao, R, Wang, S, Han, R. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition.
Mol.Cell, 81, 2021
|
|
7N3P
 
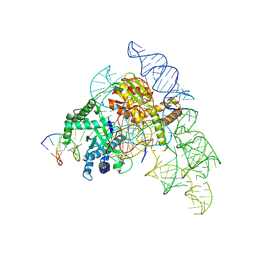 | | Cryo-EM structure of the Cas12k-sgRNA-dsDNA complex | | Descriptor: | Cas12k, DNA (5'-D(*CP*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*AP*AP*CP*CP*GP*AP*GP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*CP*TP*CP*GP*GP*TP*T)-3'), ... | | Authors: | Chang, L, Li, Z, Xiao, R, Wang, S, Han, R. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition.
Mol.Cell, 81, 2021
|
|
9BW1
 
 | | TnsABCD-DNA transpososome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Integrase, LE_polyA, ... | | Authors: | Chang, L, Wang, S. | | Deposit date: | 2024-05-20 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structure of TnsABCD transpososome reveals mechanisms of targeted DNA transposition.
Cell, 2024
|
|
6LIR
 
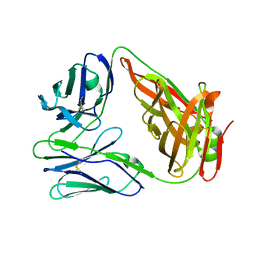 | | crystal structure of chicken TCR for 2.0 | | Descriptor: | TCR alpha chain, TCR beta chain | | Authors: | Zhang, L, Xia, C. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural and Biophysical Insights into the TCR alpha beta Complex in Chickens.
Iscience, 23, 2020
|
|
7N7V
 
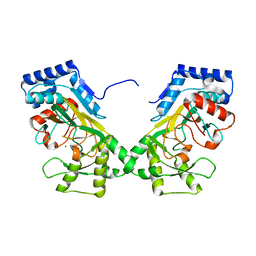 | | Crystal structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes at 2 A. | | Descriptor: | CHLORIDE ION, FE (II) ION, Predicted hydroxylase | | Authors: | Han, L, Xu, W, Ma, M, Miller, M.D, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2021-06-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes.
To Be Published
|
|
8ZAV
 
 | | alcohol dehydrogenases KpADH mutant - S9Y/F161K | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Ni, Y, Xu, G.C. | | Deposit date: | 2024-04-25 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering alcohol dehydrogenases KpADH for enhanced organic-solvent tolerance and its molecular mechanisms
To Be Published
|
|
5VFA
 
 | | RitR Mutant - C128D | | Descriptor: | Response regulator | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2017-04-07 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | RitR is an archetype for a novel family of redox sensors in the streptococci that has evolved from two-component response regulators and is required for pneumococcal colonization.
PLoS Pathog., 14, 2018
|
|
5U8M
 
 | |
5BK7
 
 | | The structure of MppP E15A mutant soaked with the substrate L-arginine | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-27 | | Release date: | 2018-09-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
7YSE
 
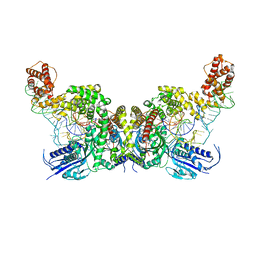 | | Crystal structure of E. coli heterotetrameric GlyRS in complex with tRNA | | Descriptor: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit, MAGNESIUM ION, ... | | Authors: | Han, L, Ju, Y, Zhou, H. | | Deposit date: | 2022-08-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | The binding mode of orphan glycyl-tRNA synthetase with tRNA supports the synthetase classification and reveals large domain movements.
Sci Adv, 9, 2023
|
|
