2E3O
 
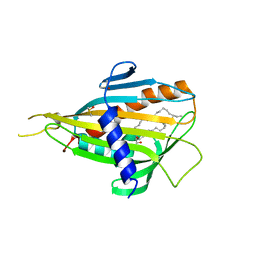 | | Crystal structure of CERT START domain in complex with C16-ceramide (P212121) | | Descriptor: | DIMETHYL SULFOXIDE, Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7XGR
 
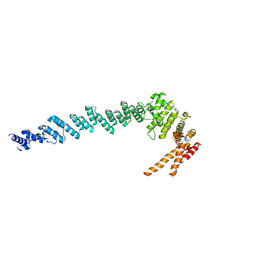 | |
7XDT
 
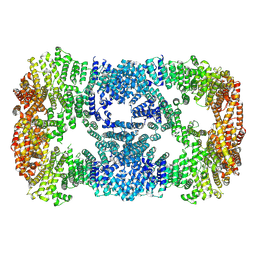 | |
5BP9
 
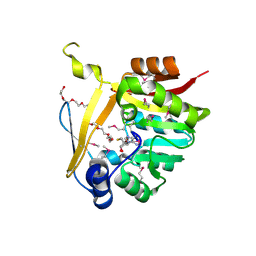 | | Crystal structure of SAM-dependent methyltransferase from Bacteroides fragilis in complex with S-Adenosyl-L-homocysteine | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Putative methyltransferase protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Cymborowski, M.T, Mason, D.V, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase fromBacteroides fragilis in complex with S-Adenosyl-L-homocysteine
to be published
|
|
4HND
 
 | | Crystal structure of the catalytic domain of Selenomethionine substituted human PI4KIIalpha in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | Deposit date: | 2012-10-19 | | Release date: | 2014-04-09 | | Last modified: | 2016-12-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
2B2X
 
 | | VLA1 RdeltaH I-domain complexed with a quadruple mutant of the AQC2 Fab | | Descriptor: | Antibody AQC2 Fab, Integrin alpha-1, MAGNESIUM ION | | Authors: | Clark, L.A, Boriack-Sjodin, P.A, Eldredge, J, Fitch, C, Friedman, B, Hanf, K.J, Jarpe, M, Liparoto, S.F, Li, Y, Lugovskoy, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Affinity enhancement of an in vivo matured therapeutic antibody using structure-based computational design
Protein Sci., 15, 2006
|
|
5BQF
 
 | | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L(+)-tartaric acid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Langner, K.M, Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Stead, M, Hillerich, B.S, Chowdhury, S, Hammonds, J, Zimmerman, M.D, Al Obaidi, N, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L-tartaric acid
to be published
|
|
4DS6
 
 | | Crystal structure of a group II intron in the pre-catalytic state | | Descriptor: | MAGNESIUM ION, Mutant Group IIC Intron | | Authors: | Chan, R.T, Robart, A.R, Rajashankar, K.R, Pyle, A.M, Toor, N. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.644 Å) | | Cite: | Crystal structure of a group II intron in the pre-catalytic state.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3G6L
 
 | |
5XG5
 
 | | Crystal structure of Mitsuba-1 with bound NAcGal | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MITSUBA-1 | | Authors: | Terada, D, Voet, A.R.D, Kamata, K, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Computational design of a symmetrical beta-trefoil lectin with cancer cell binding activity.
Sci Rep, 7, 2017
|
|
3G6M
 
 | | crystal structure of a chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with a potent inhibitor caffeine | | Descriptor: | CAFFEINE, Chitinase | | Authors: | Gan, Z, Yang, J, Lou, Z, Rao, Z, Zhang, K.-Q. | | Deposit date: | 2009-02-06 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure and mutagenesis analysis of chitinase CrChi1 from the nematophagous fungus Clonostachys rosea in complex with the inhibitor caffeine
Microbiology, 156, 2010
|
|
5EZM
 
 | | Crystal Structure of ArnT from Cupriavidus metallidurans in the apo state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferases of PMT family, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
4F1P
 
 | | Crystal Structure of mutant S554D for ArfGAP and ANK repeat domain of ACAP1 | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | Authors: | Sun, F, Pang, X, Zhang, K, Ma, J, Zhou, Q. | | Deposit date: | 2012-05-07 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
2B8O
 
 | |
1YX7
 
 | | NMR structure of Calsensin, energy minimized average structure. | | Descriptor: | Calsensin | | Authors: | Venkitaramani, D.V, Fulton, D.B, Andreotti, A.H, Johansen, K.M, Johansen, J. | | Deposit date: | 2005-02-19 | | Release date: | 2005-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of Calsensin, an invertebrate neuronal calcium-binding protein.
Protein Sci., 14, 2005
|
|
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | Descriptor: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
4HNE
 
 | | Crystal structure of the catalytic domain of human type II alpha Phosphatidylinositol 4-kinase (PI4KIIalpha) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | Authors: | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | Deposit date: | 2012-10-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
2E3Q
 
 | | Crystal structure of CERT START domain in complex with C18-ceramide (P212121) | | Descriptor: | DIMETHYL SULFOXIDE, Lipid-transfer protein CERT, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3ZEE
 
 | | Electron cyro-microscopy helical reconstruction of Par-3 N terminal domain | | Descriptor: | PARTITIONING DEFECTIVE 3 HOMOLOG | | Authors: | Zhang, Y, Wang, W, Chen, J, Zhang, K, Gao, F, Gong, W, Zhang, M, Sun, F, Feng, W. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Intrinsic Self-Assembly of Par-3 N-Terminal Domain.
Structure, 21, 2013
|
|
4NPH
 
 | | Crystal structure of SsaN from Salmonella enterica | | Descriptor: | Probable secretion system apparatus ATP synthase SsaN | | Authors: | Sugiman-Marangos, S.N, Zhang, K, Cooper, C.A, Coombes, B.K, Junop, M.S. | | Deposit date: | 2013-11-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of SsaN from Salmonella enterica
J.Biol.Chem., 2014
|
|
5C5H
 
 | | R195K E. coli MenE with bound OSB-AMS | | Descriptor: | 2-succinylbenzoate--CoA ligase, 5'-O-{[4-(2-carboxyphenyl)-4-oxobutanoyl]sulfamoyl}adenosine, MAGNESIUM ION | | Authors: | Matarlo, J.S, Shek, R, Rajashankar, K.R, Tonge, P.J, French, J.B. | | Deposit date: | 2015-06-19 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Mechanism of MenE Inhibition by Acyl-Adenylate Analogues and Discovery of Novel Antibacterial Agents.
Biochemistry, 54, 2015
|
|
3KUK
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 2'-DEOXYURIDINE, SULFATE ION, Uridine phosphorylase | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-27 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3KVY
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 1,4-anhydro-D-erythro-pent-1-enitol, SULFATE ION, URACIL, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
5C7H
 
 | | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH | | Descriptor: | Aldo-keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-24 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH
to be published
|
|
7COZ
 
 | | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase in complex with LSB-41 | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-(2-methyl-4-thiophen-2-yl-1,3-thiazol-5-yl)propanoyl]piperidine-4-carboxamide, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Dileep, K.V, Ihara, K, Sakai, N, Shirozu, M, Zhang, K.Y.J. | | Deposit date: | 2020-08-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Piperidine-4-carboxamide as a new scaffold for designing secretory glutaminyl cyclase inhibitors.
Int.J.Biol.Macromol., 170, 2020
|
|
