3FE1
 
 | | Crystal structure of the human 70kDa heat shock protein 6 (Hsp70B') ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 6, ... | | Authors: | Wisniewska, M, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-27 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
4QT4
 
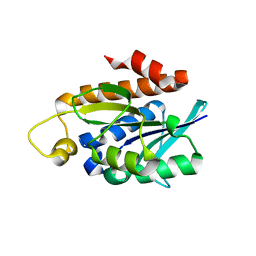 | | Crystal structure of Peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 Angstrom resolution shows the Closed Structure of the Substrate Binding Cleft | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 angstrom resolution shows the closed structure of the substrate-binding cleft.
FEBS Open Bio, 4, 2014
|
|
3OSH
 
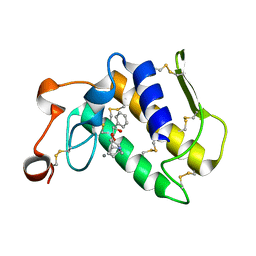 | | Crystal Structure of The Complex of Group 1 Phospholipase A2 With Atropin At 1.5 A Resolution | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Shukla, P.K, Kaushik, S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-09-09 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of The Complex of Group 1 Phospholipase A2 With Atropin At 1.5 A Resolution
To be Published
|
|
5FOJ
 
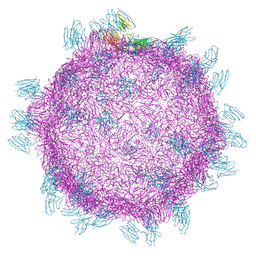 | | Cryo electron microscopy structure of Grapevine Fanleaf Virus complex with Nanobody | | Descriptor: | Nanobody, RNA2 polyprotein | | Authors: | Orlov, I, Hemmer, C, Ackerer, L, Lorber, B, Ghannam, A, Poignavent, V, Hleibieh, K, Sauter, C, Schmitt-Keichinger, C, Belval, L, Hily, J.M, Marmonier, A, Komar, V, Gersch, S, Schellenberger, P, Bron, P, Vigne, E, Muyldermans, S, Lemaire, O, Demangeat, G, Ritzenthaler, C, Klaholz, B.P. | | Deposit date: | 2015-11-22 | | Release date: | 2016-01-20 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of nanobody recognition of grapevine fanleaf virus and of virus resistance loss.
Proc.Natl.Acad.Sci.USA, 2020
|
|
6V16
 
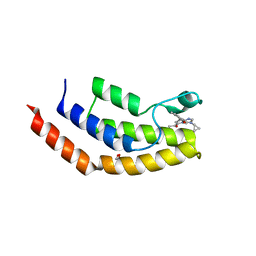 | | Crystal structure of the bromodomain of human BRD7 bound to TP472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 7, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V14
 
 | | Crystal structure of the bromodomain of human BRD9 bound to TP472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 9 | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
5J9G
 
 | |
3NAK
 
 | | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite at 3.3 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vikram, G, Singh, R.P, Singh, A.K, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite at 3.3 A resolution
To be Published
|
|
2WGH
 
 | | Human Ribonucleotide reductase R1 subunit (RRM1) in complex with dATP and Mg. | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Welin, R.M, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kragh-Nielsen, T, Kotzsch, A, Kotenyova, T, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Wisniewska, M, Nordlund, P. | | Deposit date: | 2009-04-19 | | Release date: | 2009-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Allosteric Regulation of Human Ribonucleotide Reductase by Nucleotide-Induced Oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2WNS
 
 | | Human Orotate phosphoribosyltransferase (OPRTase) domain of Uridine 5' -monophosphate synthase (UMPS) in complex with its substrate orotidine 5'-monophosphate (OMP) | | Descriptor: | CHLORIDE ION, OROTATE PHOSPHORIBOSYLTRANSFERASE, OROTIDINE-5'-MONOPHOSPHATE | | Authors: | Moche, M, Roos, A, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotyenova, T, Kotzch, A, Nielsen, T.K, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, VanDenBerg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P. | | Deposit date: | 2009-07-19 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Orotate Phosphoribosyltransferase (Oprtase) Domain of Uridine 5'-Monophosphate Synthase (Umps) in Complex with its Substrate Orotidine 5'-Monophosphate (Omp)
To be Published
|
|
6V0Q
 
 | | Crystal structure of the bromodomain of human BRD7 bound to TG003 | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 7, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V0S
 
 | |
6V0X
 
 | | Crystal structure of the bromodomain of human BRD9 bound to sunitinib | | Descriptor: | Bromodomain-containing protein 9, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Karim, M.R, Chan, A, Zhu, J.Y, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1B
 
 | | Crystal structure of the bromodomain of human BRD9 bound to I-BRD9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 9, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6UZF
 
 | | Crystal structure of the unliganded bromodomain of human BRD9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 9, DIMETHYL SULFOXIDE, ... | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V17
 
 | | Crystal structure of the bromodomain of human BRD7 bound to I-BRD9 | | Descriptor: | Bromodomain-containing protein 7, CHLORIDE ION, N'-[1,1-bis(oxidanylidene)thian-4-yl]-5-ethyl-4-oxidanylidene-7-[3-(trifluoromethyl)phenyl]thieno[3,2-c]pyridine-2-carboximidamide | | Authors: | Karim, M.R, Chan, A, Schonbrunn, E. | | Deposit date: | 2019-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
5UJC
 
 | | Crystal structure of a C.elegans B12-trafficking protein CblC, a human MMACHC homologue | | Descriptor: | CO-METHYLCOBALAMIN, D(-)-TARTARIC ACID, GLYCEROL, ... | | Authors: | Li, Z, Shanmuganathan, A, Ruetz, M, Yamada, K, Lesniak, N.A, Krautler, B, Brunold, T.C, Banerjee, R, Koutmos, M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Coordination chemistry controls the thiol oxidase activity of the B12-trafficking protein CblC.
J. Biol. Chem., 292, 2017
|
|
4PTM
 
 | | Crystal Structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine, a hydrolyzed product of hexasaccharide at 1.7 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Madhuprakash, J, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Podile, A.R, Singh, T.P. | | Deposit date: | 2014-03-11 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine, a hydrolyzed product of hexasaccharide at 1.7 Angstrom resolution
To be Published
|
|
5TSH
 
 | | PilB from Geobacter metallireducens bound to AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | McCallum, M, Tammam, S, Khan, A, Burrows, L, Howell, P.L. | | Deposit date: | 2016-10-28 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular mechanism of the type IVa pilus motors.
Nat Commun, 8, 2017
|
|
7N99
 
 | | SDE2 SAP domain apo structure | | Descriptor: | Isoform 2 of Replication stress response regulator SDE2 | | Authors: | Paung, Y, Weinheimer, A.S, Rageul, J, Khan, A, Ho, B, Tong, M, Alphonse, S, Seeliger, M.A, Kim, H. | | Deposit date: | 2021-06-17 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Extended DNA-binding interfaces beyond the canonical SAP domain contribute to the function of replication stress regulator SDE2 at DNA replication forks.
J.Biol.Chem., 298, 2022
|
|
3DKP
 
 | | Human DEAD-box RNA-helicase DDX52, conserved domain I in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lehtio, L, Karlberg, T, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, van den Berg, S, Welin, M, Wisniewska, M, Wikstrom, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
2WWP
 
 | | Crystal structure of the human lipocalin-type prostaglandin D synthase | | Descriptor: | CHLORIDE ION, PROSTAGLANDIN-H2 D-ISOMERASE, THIOCYANATE ION | | Authors: | Roos, A.K, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotyenova, T, Kotzch, A, Kraulis, P, Markova, N, Moche, M, Nielsen, T.K, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Dynamic Insights Into Substrate Binding and Catalysis of Human Lipocalin Prostaglandin D Synthase.
J.Lipid Res., 54, 2013
|
|
3FO5
 
 | | Human START domain of Acyl-coenzyme A thioesterase 11 (ACOT11) | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van-Den-Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Shueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-27 | | Release date: | 2009-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural analysis of lipid binding START domains.
Plos One, 6, 2011
|
|
3FE2
 
 | | Human DEAD-BOX RNA helicase DDX5 (P68), conserved domain I in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX5, ... | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-27 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases
Plos One, 5, 2010
|
|
6HMA
 
 | | Improved model derived from cryo-EM map of Staphylococcus aureus large ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Eyal, Z, Cimicata, G, Matzov, D, Fox, T, de Val, N, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Improved model derived from cryo-EM map of Staphylococcus aureus large ribosomal subunit
To Be Published
|
|
